Unveiling the Structure of Heart Rate Variability (HRV) Indices: A Data-driven Meta-clustering Approach
Supplementary Materials
Introduction
Heart Rate Variability (HRV) can be estimated using a myriad of mathematical indices, but the lack of systematic comparison between these indices renders the interpretation and evaluation of results tedious. In this study, we assessed the relationship between 57 HRV metrics collected from 302 human recordings using a variety of structure-analysis algorithms. We then applied a meta-clustering approach that combines their results to obtain a robust and reliable view of the observed relationships. We found that HRV metrics can be clustered into 3 groups, representing the distribution-related features, harmony-related features and frequency/complexity features. From there, we described and discussed their associations, and derived recommendations on which indices to prioritize for parsimonious, yet comprehensive HRV-related data analysis and reporting.
Heart Rate Variability (HRV), reflecting the heart’s ability to effectively regulate and adapt to internal and external environmental changes, has been linked with many physical and mental health outcomes Forte et al. (2019). Conventionally, the various indices used in the assessment of HRV were broadly categorized based on the nature of their mathematical approach, in categories including time-domain, frequency-domain, and nonlinear dynamics.
Time-domain analysis presents the simplest and most straightforward method of quantifying HRV from the original normal (i.e., excluding abnormal beats such as ectopic beats) heartbeat intervals or NN intervals. Some commonly derived indices include SDNN, the standard deviation of all NN intervals, RMSSD, the root mean square of the sum of successive differences of NN intervals, and pNN50, the percentage of adjacent NN intervals separated by more than 50ms. While time-domain methods offers computational ease, they are unable to distinguish between the contributions of sympathetic and parasympathetic branches. Frequency-domain analysis, on the other hand, targets the assessment of these different regulatory mechanisms by investigating how the HRV power spectrum distributes across different frequency bands (e.g, low frequency, LF or high frequency, HF). Other indices that fall under the frequency domain include derivatives of the aforementioned components, such as the ratio of LF to HF (LF/HF) power and their normalized (e.g., LFn, HFn) and natural logarithmic variants (e.g., LnHF). Finally, drawn from concepts of non-linear dynamics and chaos theory (Golberger, 1996), non-linear analysis was later introduced to better characterize the complex physiological mechanisms underlying HRV regulation. Prominent indices include measures obtained from a Poincaré plot where an ellipse is fitted to a scatterplot of each NN interval against its preceding one (e.g., the standard deviation of the short-term, SD1 and long-term, SD2 NN interval variability, as well as its corresponding ratio, SD1/SD2, Brennan et al., 2001). Other non-linear indices that fall under this category, such as Detrended Fluctuation Analysis (DFA), multi-fractal DFA (MF-DFA) and correlation dimension (CD), account for the fractal properties of HRV, while entropy measures like approximate entropy (ApEn), sample entropy (SampEn), and multiscale entropy (MSE) quantify the amount of regularity in the HR time series (Voss et al., 2009). For a more comprehensive description of all HRV indices, see Pham et al. (2021).
Despite the rising popularity of HRV analysis as a real-time, noninvasive technique for investigating health and disease, there are some shared similarities (and even overlaps) between the multitude of HRV indices that are not yet well understood. Early studies have investigated the relationships between time-domain and frequency-domain indices, showing that not only were RMSSD and pNN50 strongly correlated with each other (above 0.9, Bigger Jr et al., 1989), they were also highly associated with HF power (Bigger Jr et al., 1989; Kleiger et al., 2005; Otzenberger et al., 1998), suggesting that these measures could be treated as surrogates for each other in assessing the parasympathetic modulation of HRV. This observation is warranted given that the former is computed from the differences across consecutive NN intervals, and hence, they reflect mainly high frequency oscillatory patterns in HR and are independent of long-term changes. On the other hand, SDNN, which has been thought to reflect both sympathetic and parasympathetic activity, is correlated to total power (TP) in HRV power spectrum (Bigger Jr et al., 1989). Recent years also witnessed the emergence of debates regarding the traditional conceptualization of SD1 and SD2 as non-linear indices, particularly when Ciccone et al. (2017) proposed that RMSSD and SD1 were mathematically equivalent. Many studies that report both of these short-term HRV indices independently often arrive at identical statistical results without addressing this equivalence Leite et al. (2015). Additionally, other studies have also drawn similarities between SD1/SD2 and LF/HF in their indexing of the balance between short- and long-term HRV (Brennan et al., 2002; Guzik et al., 2007).
Overall, there is a need for a greater data-driven understanding of the relationship between the multitude of HRV indices and their respective groupings. While there exist different approaches to assign data to different groups based on their level of associations (see Nguyen Phuc Thu et al., 2019), there is no gold standard or clear guidelines to determine the most appropriate method for grouping of physiological indices. As such, choosing one method and presenting its solution as a definitive one can be uninformative or even misleading. The aim of this study is to explore the structure of HRV indices by using a consensus-based methodology (Bhattacharjee et al., 2001; Kuncheva, 2014; Monti et al., 2003), hereafter referred to as meta-clustering, where the results of multiple structure analysis approaches are systematically combined to highlight the most robust associations between the indices.
Methods
In total, the electrocardiogram (ECG) data of 302 participants were extracted from 6 databases. The raw data as well as the entire analysis script (including details and additional analyses) can be found at this GitHub repository (https://github.com/Tam-Pham/HRVStructure).
Databases
The Glasgow University Database (GUDB) database (Howell & Porr, 2018) contains ECG recordings from 25 healthy participants (> 18 years old) performing five different two-minute tasks (sitting, doing a maths test on a tablet, walking on a treadmill, running on a treadmill, using a handbike). All recordings were sampled at 250 Hz.
The MIT-BIH Arrhythmia Database (MIT-Arrhythmia and MIT-Arrhythmia-x) database (Moody & Mark, 2001) contains 48 ECG recordings (25 men, 32-89 years old; 22 women, 23-89 years old) from a mixed population of patients. All recordings were sampled at 360 Hz and lasted for 30 minutes.
The Fantasia database (Iyengar et al., 1996) contains ECG recordings from 20 young (21-34 years old) and 20 elderly (68-85 years old) healthy participants. All participants remained in a resting state in sinus rhythm while watching the movie Fantasia (Disney, 1940) that helped to maintain wakefulness. All recordings were sampled at 250 Hz and lasted for 120 minutes.
The MIT-BIH Normal Sinus Rhythm Database (MIT-Normal) database (Goldberger et al., 2000) contains long-term (\(\approx\). 24h) ECG recordings from 18 participants (5 men, 26-45 years old; 13 women, 20-50 years old). All recordings were sampled at 128 Hz and due to memory limits, we kept only the second and third hours of each recording (with the loose assumption that the first hour might be less representative of the rest of the recording and a duration of 120 minutes to match those from Fantasia database).
The MIT-BIH Long-term ECG Database (MIT-Long-term) database (Goldberger et al., 2000) contains long-term (14 to 22 hours each) ECG recordings from 7 participants (6 men, 46-88 years old; 1 woman, 71 years old). All recordings were sampled at 128 Hz and due to memory limits, we kept only the second and third hours of each recording.
The last dataset came from resting-state https://github.com/neuropsychology/RestingState recordings of authors’ other empirical studies. This dataset contains ECG recordings sampled at 4000 Hz from 43 healthy participants (> 18 years old) that underwent 8 minutes of eyes-closed, seated position, resting state.
Data Processing
The default processing pipeline of NeuroKit2 (Makowski et al., 2021) was used for cleaning and processing of raw ECG recordings as well as for the computation of all HRV indices. Note that except for the resting-state dataset, data from online database was not in the form of ECG recordings but sample locations of annotated heartbeats (R-peaks).
Data Analysis
One of the core “issues” of statistical clustering is that, in many cases, different methods will give different results. The meta-clustering approach proposed by easystats (that finds echoes in consensus clustering; see Monti et al., 2003) consists of treating the unique clustering solutions as a ensemble, from which we can derive a probability matrix. This matrix contains, for each pair of observations, the probability of being in the same cluster. For instance, if the 6th and the 9th row of a dataframe has been assigned to a similar cluster by 5 out of 10 clustering methods, then its probability of being grouped together is 0.5. Essentially, meta-clustering approach is based on the hypothesis that, as each clustering algorithm embodies a different prism by which it sees the data, running an infinite amount of algorithms would result in the emergence of the “true” clusters. As the number of algorithms and parameters is finite, the probabilistic perspective is a useful proxy. This method is interesting where there is no obvious reasons to prefer one over another clustering method, as well as to investigate how robust some clusters are under different algorithms.
In this analysis, we first correlated the HRV indices (using the correlations function from the correlation package, see Makowski et al., 2020) and removed indices that were perfectly correlated with each other, before removing outliers based on the median absolute deviation from the median [see the check_outliers function in the performance R package; (ludecke2019performance?)]. Following the meta-clustering approach, multiple clustering methods were used to analyze the associations between the HRV indices, namely, factor analysis, k-means clustering, k-meloids clustering, hierarchical clustering, density-based spatial clustering of applications with noise (DBSCAN), hierarchical density-based spatial clustering of applications with noise (HBSCAN), mixture model approach, and exploratory graph analysis (EGA). We then jointly considered the results obtained from these multiple methods by estimating the probability for each pair of indices to be clustered together. Ultimately, the analysis aimed to to provide a more robust overview of the clustering results and facilitate a more thorough conceptual understanding of these patterns.
Data processing was carried out with R (R Core Team, 2019) and the easystats ecosystem (Ludecke et al., 2019; Makowski et al., 2019). The raw data, as well as the full reproducible analysis script, including additional description of each approach and the solutions of each individual clustering method, are available at this GitHub repository (https://github.com/Tam-Pham/HRVStructure).
Results
library(tidyverse)
library(easystats)
data <- read.csv("data/data_combined.csv", stringsAsFactors = FALSE) %>%
select(-HRV_ULF, -HRV_VLF) %>% # insufficient recording length to compute
select(-HRV_SDANN1, -HRV_SDANN2, -HRV_SDANN5,
-HRV_SDNNI1, -HRV_SDNNI2, -HRV_SDNNI5) %>% # insufficient recording length to compute
setNames(stringr::str_remove(names(.), "HRV_")) %>%
mutate_all(function(x) {
x[is.infinite(x)] <- NA
return(x) })cat("This study includes",
nrow(data),
"participants from",
length(unique(data$Database)),
"databases.")This study includes 302 participants from 11 databases.
Preprocessing
Convenience Functions
library(ggraph)Cleaning
clean_names <- function(n) {
n[n == "DFA_alpha1_ExpMean"] <- "DFA α1 ExpMean"
n[n == "DFA_alpha2_ExpMean"] <- "DFA α2 ExpMean"
n[n == "DFA_alpha1_ExpRange"] <- "DFA α1 ExpRange"
n[n == "DFA_alpha2_ExpRange"] <- "DFA α2 ExpRange"
n[n == "DFA_alpha1_DimMean"] <- "DFA α1 DimMean"
n[n == "DFA_alpha2_DimMean"] <- "DFA α2 DimMean"
n[n == "DFA_alpha1_DimRange"] <- "DFA α1 DimRange"
n[n == "DFA_alpha2_DimRange"] <- "DFA α2 DimRange"
n[n == "DFA_alpha1"] <- "DFA α1"
n[n == "DFA_alpha2"] <- "DFA α2"
n[n == "CSI_Modified"] <- "CSI (modified)"
n
}
assign_clusters <- function(table_clusters, data, clusters, col = "Hclust") {
table_clusters[[col]] <- NA
table_clusters[[col]][table_clusters$Index %in% row.names(data)] <- clusters
table_clusters[[col]][table_clusters[[col]] == "0"] <- seq(length(unique(clusters)),sum(clusters == "0") + length(unique(clusters)) - 1)
table_clusters
}Dendrogram
plot_dendrogram <- function(model, clusters = NULL, points = TRUE, dis = NULL, ylim = -0.3, nudge_y = -0.02, angle = 90, h = NULL, edges = "elbow") {
if(is.null(clusters)) {
data <- tidygraph::as_tbl_graph(model)
} else {
data <- tidygraph::as_tbl_graph(model) %>%
tidygraph::activate(nodes) %>%
mutate(cluster = as.character(clusters[label]))
}
if(!is.null(dis)) {
data <- data %>%
mutate(dis = dis[label])
}
# Tree
p <- ggraph(data, layout = 'dendrogram', height = height)
if(edges == "elbow") {
p <- p + geom_edge_elbow()
} else {
p <- p + geom_edge_diagonal()
}
# Hline
if(!is.null(h)) p <- p + geom_hline(yintercept = h, linetype = "dotted")
# Points
if(points) {
if(is.null(clusters)) {
p <- p + geom_node_point(aes(filter=leaf))
} else {
p <- p + geom_node_point(aes(color = cluster, filter = leaf, size = dis))
}
}
if(is.null(clusters)) {
p <- p + geom_node_text(aes(label=label), angle = angle, hjust=1, nudge_y = nudge_y)
} else {
p <- p + geom_node_text(aes(label=label, color = cluster), angle = angle, hjust=1, nudge_y = nudge_y) +
scale_colour_material_d(palette = "rainbow", guide = "none", na.value = "grey")
}
p +
ylim(ylim, NA) +
theme_void()
}Correlations
plot_correlation <- function(data, x = "Recording_Length", y = "RMSSD", xlab = x, ylab = y, color = "red", label = NULL) {
data %>%
ggplot(aes_string(x = x, y = y)) +
geom_point2(size = 4, alpha = 0.66) +
geom_smooth(method = "lm", formula = y ~ poly(x, 2), color = color, fill = color, alpha = 0.1, size = 1.5, se = FALSE) +
annotate("label",
x = min(data[[x]], na.rm = TRUE),
y = max(data[[y]], na.rm = TRUE),
label = label,
hjust = 0
) +
xlab(xlab) +
ylab(ylab) +
theme_modern() +
ggside::geom_xsidedensity() +
ggside::geom_ysidedensity() +
ggside::scale_xsidey_continuous(breaks = NULL, labels = "", expand = expansion(c(0, 0))) +
ggside::scale_ysidex_continuous(breaks = NULL, labels = "", expand = expansion(c(0, 0)))
}
plot_correlations <- function(data, r, color = NA) {
r$label <- paste0(
"r = ",
insight::format_value(r$r),
insight::format_p(r$p, stars_only = TRUE),
", ",
insight::format_ci(r$CI_low, r$CI_high, r$CI, zap_small = TRUE)
)
r$xlab <- clean_names(r$Parameter1)
r$ylab <- clean_names(r$Parameter2)
ps <- list()
for(i in 1:nrow(r)) {
ps[[i]] <- plot_correlation(data,
x = r$Parameter1[i],
y = r$Parameter2[i],
xlab = r$xlab[i],
ylab = r$ylab[i],
color = "red",
label = r$label[i]) +
theme(axis.title.x = element_blank())
}
ggpubr::ggarrange(plotlist = ps)
}Indices Selection
Remove Equivalent
data %>%
select(-Participant, -Recording_Length, -Database) %>%
correlation::correlation() %>%
filter(abs(r) > 0.999) %>%
arrange(Parameter1, desc(abs(r))) %>%
format()> Parameter1 Parameter2 r 95% CI t(300) p
> 1 C1d C1a -1.00 [-1.00, -1.00] -Inf < .001***
> 2 C2d C2a -1.00 [-1.00, -1.00] -1.16e+09 < .001***
> 3 Cd Ca -1.00 [-1.00, -1.00] -Inf < .001***
> 4 RMSSD SDSD 1.00 [ 1.00, 1.00] 54568.34 < .001***
> 5 RMSSD SD1 1.00 [ 1.00, 1.00] 54568.34 < .001***
> 6 RMSSD SD1d 1.00 [ 1.00, 1.00] 594.68 < .001***
> 7 RMSSD SD1a 1.00 [ 1.00, 1.00] 519.33 < .001***
> 8 SD1 SD1d 1.00 [ 1.00, 1.00] 595.51 < .001***
> 9 SD1 SD1a 1.00 [ 1.00, 1.00] 518.73 < .001***
> 10 SDNN SDNNa 1.00 [ 1.00, 1.00] 756.47 < .001***
> 11 SDNN SDNNd 1.00 [ 1.00, 1.00] 617.85 < .001***
> 12 SDSD SD1 1.00 [ 1.00, 1.00] 8.22e+08 < .001***
> 13 SDSD SD1d 1.00 [ 1.00, 1.00] 595.51 < .001***
> 14 SDSD SD1a 1.00 [ 1.00, 1.00] 518.73 < .001***data <- data %>%
select(-SDSD, -SD1, -SD1d, -SD1a, -CVSD) %>% # Same as RMSSD
select(-SDNNd, -SDNNa) %>% # Same as SDNN
select(-SD2d, -SD2a) %>% # Same as SD2
select(-Cd) %>% # Same as Ca
select(-C1d, -C2d) # Same as C1a and C2a
hrv_cols <- names(select(data, -Participant, -Recording_Length, -Database))Outliers Removal
Before Removal
data[hrv_cols] %>%
normalize() %>%
estimate_density() %>%
plot() +
facet_wrap(~Parameter, scales = "free") +
theme(legend.position = "none",
axis.title.x = element_blank(),
axis.title.y = element_blank(),
axis.text.y = element_blank())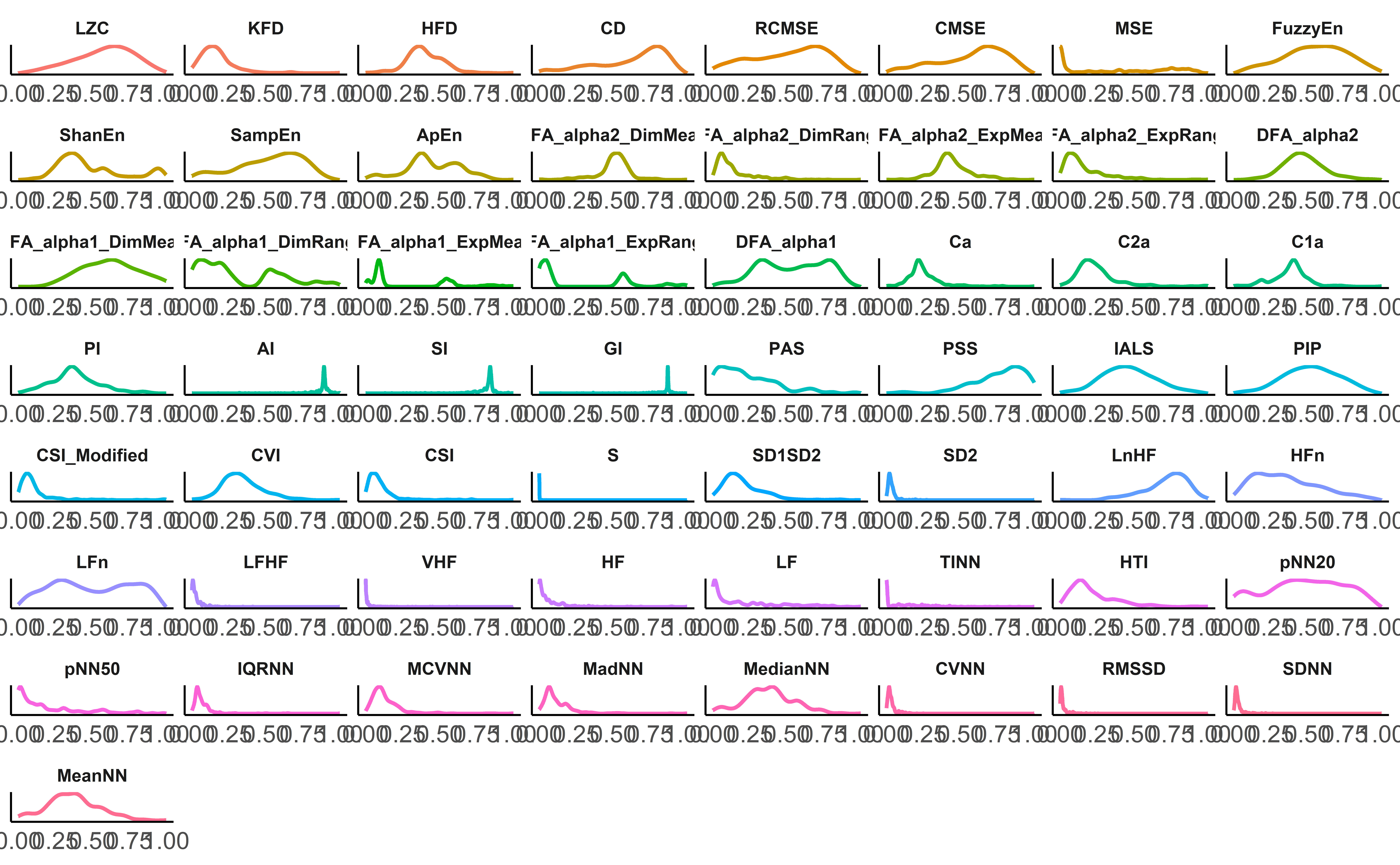
After Removal
df_outliers <- data.frame(hrv_cols = vector(mode = "character", length = length(hrv_cols)),
outliers = vector(mode = "integer", length = length(hrv_cols)))
for(i in 1:length(hrv_cols)) {
col <- hrv_cols[i]
outliers <- as.logical(performance::check_outliers(data[[col]], method = "zscore_robust", threshold = qnorm(0.9999)))
data[outliers, col] <- NA
df_outliers$hrv_cols[i] <- col
df_outliers$outliers[i] <- insight::format_value(sum(outliers) / nrow(data))
cat(paste0("\n-",
col, ": ",
sum(outliers),
" outliers (",
insight::format_value(sum(outliers) / nrow(data), as_percent = TRUE),
") detected and removed."))
}>
> -MeanNN: 1 outliers (0.33%) detected and removed.
> -SDNN: 27 outliers (8.94%) detected and removed.
> -RMSSD: 38 outliers (12.58%) detected and removed.
> -CVNN: 29 outliers (9.60%) detected and removed.
> -MedianNN: 1 outliers (0.33%) detected and removed.
> -MadNN: 13 outliers (4.30%) detected and removed.
> -MCVNN: 13 outliers (4.30%) detected and removed.
> -IQRNN: 17 outliers (5.63%) detected and removed.
> -pNN50: 11 outliers (3.64%) detected and removed.
> -pNN20: 0 outliers (0.00%) detected and removed.
> -HTI: 11 outliers (3.64%) detected and removed.
> -TINN: 9 outliers (2.98%) detected and removed.
> -LF: 22 outliers (7.28%) detected and removed.
> -HF: 25 outliers (8.28%) detected and removed.
> -VHF: 46 outliers (15.23%) detected and removed.
> -LFHF: 21 outliers (6.95%) detected and removed.
> -LFn: 0 outliers (0.00%) detected and removed.
> -HFn: 0 outliers (0.00%) detected and removed.
> -LnHF: 1 outliers (0.33%) detected and removed.
> -SD2: 19 outliers (6.29%) detected and removed.
> -SD1SD2: 13 outliers (4.30%) detected and removed.
> -S: 53 outliers (17.55%) detected and removed.
> -CSI: 17 outliers (5.63%) detected and removed.
> -CVI: 2 outliers (0.66%) detected and removed.
> -CSI_Modified: 25 outliers (8.28%) detected and removed.
> -PIP: 0 outliers (0.00%) detected and removed.
> -IALS: 0 outliers (0.00%) detected and removed.
> -PSS: 8 outliers (2.65%) detected and removed.
> -PAS: 4 outliers (1.32%) detected and removed.
> -GI: 81 outliers (26.82%) detected and removed.
> -SI: 58 outliers (19.21%) detected and removed.
> -AI: 54 outliers (17.88%) detected and removed.
> -PI: 2 outliers (0.66%) detected and removed.
> -C1a: 10 outliers (3.31%) detected and removed.
> -C2a: 15 outliers (4.97%) detected and removed.
> -Ca: 14 outliers (4.64%) detected and removed.
> -DFA_alpha1: 0 outliers (0.00%) detected and removed.
> -DFA_alpha1_ExpRange: 116 outliers (38.41%) detected and removed.
> -DFA_alpha1_ExpMean: 116 outliers (38.41%) detected and removed.
> -DFA_alpha1_DimRange: 0 outliers (0.00%) detected and removed.
> -DFA_alpha1_DimMean: 0 outliers (0.00%) detected and removed.
> -DFA_alpha2: 4 outliers (1.32%) detected and removed.
> -DFA_alpha2_ExpRange: 16 outliers (5.30%) detected and removed.
> -DFA_alpha2_ExpMean: 7 outliers (2.32%) detected and removed.
> -DFA_alpha2_DimRange: 25 outliers (8.28%) detected and removed.
> -DFA_alpha2_DimMean: 18 outliers (5.96%) detected and removed.
> -ApEn: 0 outliers (0.00%) detected and removed.
> -SampEn: 0 outliers (0.00%) detected and removed.
> -ShanEn: 0 outliers (0.00%) detected and removed.
> -FuzzyEn: 0 outliers (0.00%) detected and removed.
> -MSE: 0 outliers (0.00%) detected and removed.
> -CMSE: 0 outliers (0.00%) detected and removed.
> -RCMSE: 0 outliers (0.00%) detected and removed.
> -CD: 0 outliers (0.00%) detected and removed.
> -HFD: 2 outliers (0.66%) detected and removed.
> -KFD: 7 outliers (2.32%) detected and removed.
> -LZC: 0 outliers (0.00%) detected and removed.cat(paste0("\n-On average, " ,
insight::format_value(sum(as.double(df_outliers$outliers)) / length(hrv_cols), as_percent = TRUE),
" of data was detected as outliers and removed."))>
> -On average, 5.61% of data was detected as outliers and removed.data[hrv_cols] %>%
normalize() %>%
estimate_density() %>%
plot() +
facet_wrap(~Parameter, scales = "free") +
theme(legend.position = "none",
axis.title.x = element_blank(),
axis.title.y = element_blank(),
axis.text.y = element_blank())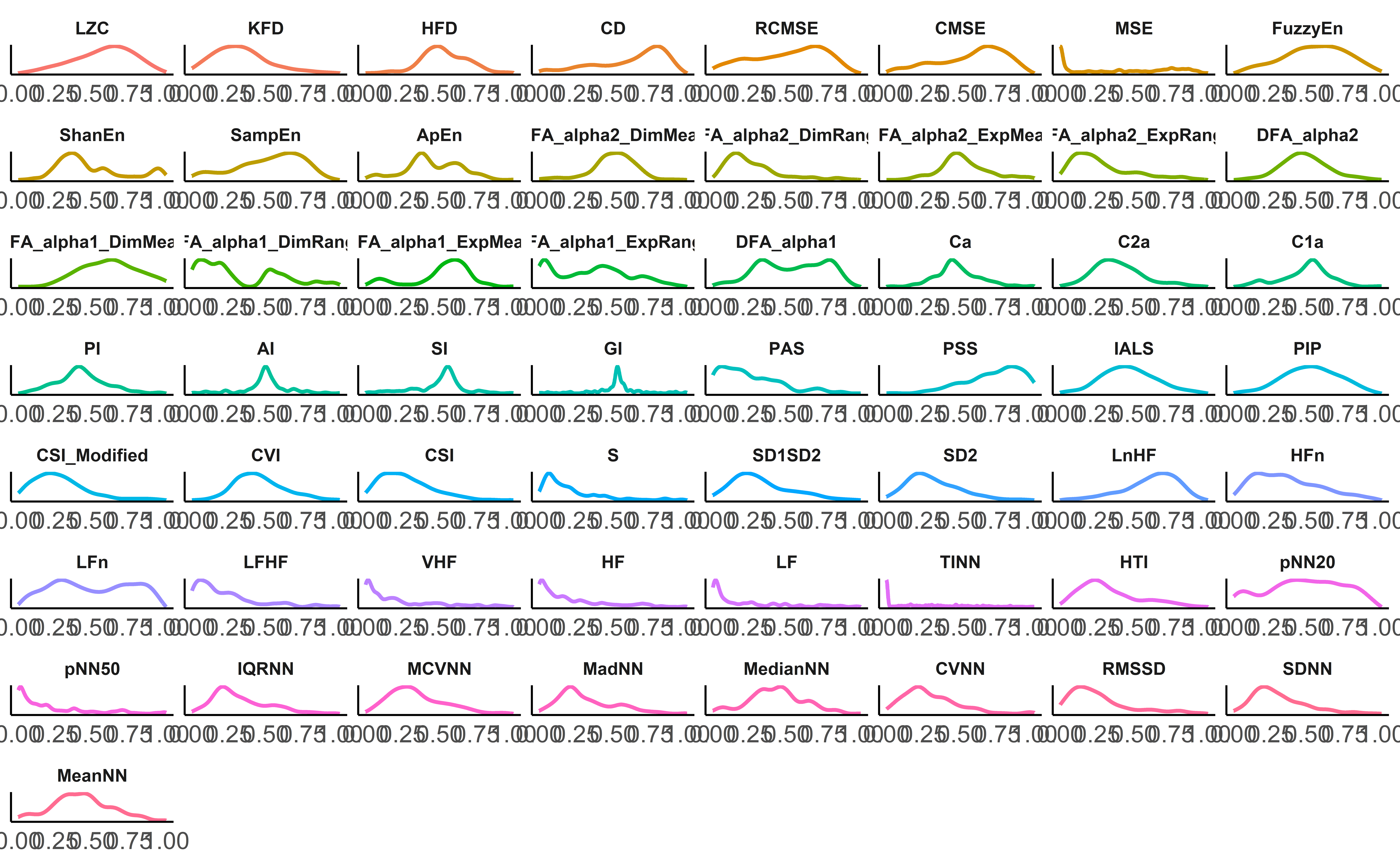
Dimensional Structure
Dimension Reduction
PCA and EFA are both dimension reduction techniques that aim to group the observed variables into smaller sets of constructs. While PCA aims to extract the sets/ components that can explain all or most of the variance present in the observed variables, EFA focuses on the variance that is shared among the variables and looks for sets/ factors that could explain the correlations among them.
Principal Component Analysis
How many components
r <- correlation::correlation(data[hrv_cols]) %>%
as.matrix() %>%
correlation::cor_smooth(verbose = FALSE)
set.seed(3)
# n_max to 28 to have at least 2 var per component
n <- parameters::n_components(data[hrv_cols], cor=r, rotation = "promax", package = "all", n_max = 28)
n> # Method Agreement Procedure:
>
> The choice of 1 dimensions is supported by 2 (15.38%) methods out of 13 (t, p).plot(n) 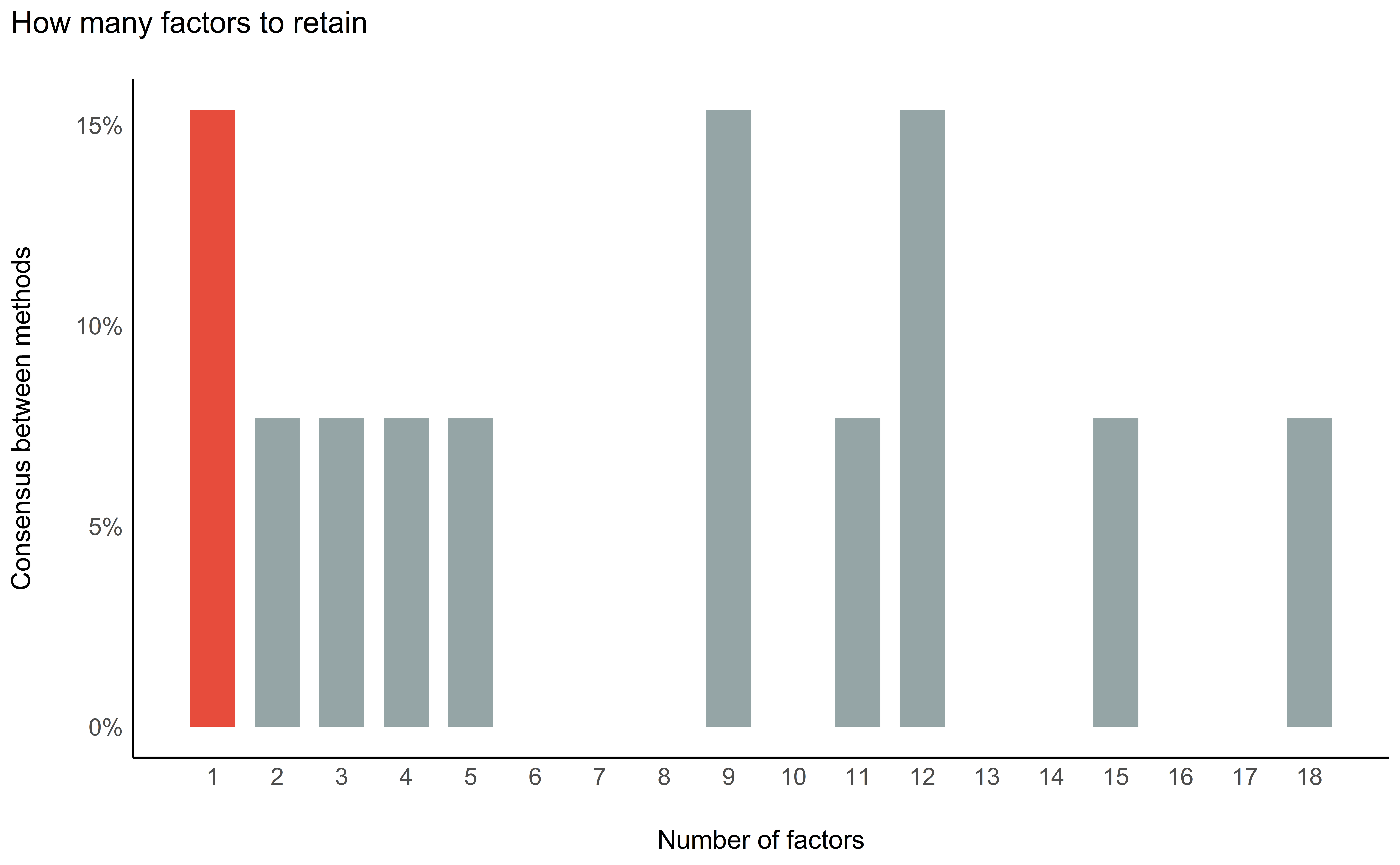
9-components
set.seed(3)
pca <- parameters::principal_components(data[hrv_cols], n=9, rotation = "promax")
knitr::kable(pca,
booktabs = TRUE,
caption = "Item loadings for PCA 9-component solutions."
)| Variable | RC1 | RC2 | RC3 | RC5 | RC7 | RC4 | RC8 | RC6 | RC9 | Complexity | Uniqueness | MSA |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MeanNN | 0.05 | 0.10 | 0.09 | 0.38 | 1.07 | -0.03 | 0.05 | -0.15 | 0.14 | 1.4 | 0.12 | 0.17 |
| SDNN | 0.91 | -0.13 | -0.07 | 0.03 | 0.11 | -0.06 | 0.02 | 0.16 | 0.03 | 1.2 | 0.02 | 0.28 |
| RMSSD | 0.57 | -0.21 | -0.56 | 0.10 | 0.15 | -0.01 | -0.04 | 0.23 | 0.02 | 2.8 | 0.05 | 0.29 |
| CVNN | 1.00 | -0.24 | -0.11 | -0.18 | -0.34 | -0.10 | 0.00 | 0.17 | -0.02 | 1.6 | 0.06 | 0.27 |
| MedianNN | 0.05 | 0.10 | 0.09 | 0.39 | 1.07 | -0.03 | 0.06 | -0.15 | 0.15 | 1.4 | 0.13 | 0.15 |
| MadNN | 0.96 | 0.12 | 0.12 | -0.04 | 0.06 | -0.06 | 0.02 | 0.09 | 0.04 | 1.1 | 0.05 | 0.24 |
| MCVNN | 1.07 | 0.03 | 0.09 | -0.25 | -0.39 | -0.10 | -0.01 | 0.09 | 0.00 | 1.4 | 0.06 | 0.31 |
| IQRNN | 0.96 | 0.14 | 0.12 | -0.03 | 0.06 | -0.08 | 0.01 | 0.11 | 0.01 | 1.1 | 0.04 | 0.24 |
| pNN50 | 0.55 | 0.47 | -0.25 | 0.16 | 0.18 | -0.01 | 0.05 | 0.21 | -0.03 | 3.2 | 0.13 | 0.25 |
| pNN20 | 0.62 | 0.55 | -0.14 | 0.01 | 0.14 | -0.15 | 0.06 | 0.02 | 0.08 | 2.4 | 0.08 | 0.25 |
| HTI | 0.94 | 0.00 | 0.16 | -0.06 | 0.10 | -0.08 | -0.03 | 0.06 | 0.03 | 1.1 | 0.04 | 0.26 |
| TINN | -0.02 | 0.16 | -0.11 | -0.05 | 0.07 | 0.08 | 0.73 | -0.05 | 0.11 | 1.3 | 0.25 | 0.47 |
| LF | -0.29 | 0.09 | 0.24 | -0.24 | 0.25 | -0.37 | 0.44 | 0.26 | -0.02 | 5.7 | 0.13 | 0.18 |
| HF | -0.11 | 0.33 | -0.35 | -0.17 | 0.04 | -0.13 | 0.35 | 0.15 | -0.17 | 4.8 | 0.29 | 0.18 |
| VHF | -0.20 | 0.24 | -0.11 | 0.12 | 0.07 | -0.13 | 0.11 | 0.32 | 0.35 | 4.8 | 0.33 | 0.46 |
| LFHF | -0.06 | -0.09 | 0.98 | 0.02 | 0.35 | -0.20 | -0.05 | 0.15 | 0.08 | 1.4 | 0.20 | 0.13 |
| LFn | -0.26 | -0.11 | 0.31 | -0.12 | -0.03 | -0.04 | 0.62 | 0.32 | 0.02 | 2.7 | 0.23 | 0.14 |
| HFn | -0.17 | 0.40 | -0.78 | -0.14 | -0.31 | 0.12 | -0.13 | -0.13 | 0.00 | 2.3 | 0.08 | 0.22 |
| LnHF | -0.16 | 0.24 | -0.52 | -0.19 | -0.06 | -0.02 | 0.42 | 0.16 | 0.04 | 3.2 | 0.07 | 0.25 |
| SD2 | 0.95 | -0.10 | 0.06 | 0.01 | 0.09 | -0.08 | 0.06 | 0.13 | 0.02 | 1.1 | 0.03 | 0.27 |
| SD1SD2 | -0.06 | -0.11 | -0.79 | 0.03 | 0.16 | -0.05 | -0.33 | 0.06 | 0.10 | 1.6 | 0.07 | 0.20 |
| S | 0.72 | -0.20 | -0.36 | 0.09 | 0.12 | 0.02 | 0.03 | 0.24 | 0.00 | 2.1 | 0.10 | 0.66 |
| CSI | -0.06 | 0.15 | 0.82 | 0.05 | -0.19 | 0.15 | 0.03 | 0.00 | -0.07 | 1.3 | 0.10 | 0.49 |
| CVI | 0.77 | -0.17 | -0.26 | -0.04 | 0.21 | -0.16 | 0.01 | 0.06 | 0.11 | 1.7 | 0.07 | 0.48 |
| CSI_Modified | 0.69 | 0.12 | 0.76 | -0.02 | -0.03 | -0.04 | -0.04 | -0.05 | 0.03 | 2.1 | 0.09 | 0.59 |
| PIP | -0.08 | -0.05 | -0.04 | 0.99 | 0.22 | 0.02 | -0.10 | -0.01 | -0.12 | 1.2 | 0.04 | 0.26 |
| IALS | -0.12 | 0.03 | -0.07 | 1.04 | 0.21 | 0.07 | -0.04 | 0.02 | -0.12 | 1.2 | 0.05 | 0.28 |
| PSS | -0.08 | 0.14 | 0.01 | 1.08 | 0.44 | 0.08 | -0.01 | -0.03 | -0.13 | 1.4 | 0.11 | 0.11 |
| PAS | -0.15 | -0.32 | 0.12 | 0.61 | -0.05 | 0.01 | -0.11 | 0.15 | -0.01 | 2.0 | 0.19 | 0.19 |
| GI | -0.29 | -0.13 | -0.07 | 0.02 | -0.12 | 0.98 | -0.04 | 0.26 | 0.13 | 1.5 | 0.16 | 0.15 |
| SI | -0.34 | -0.12 | -0.04 | -0.09 | -0.15 | 0.86 | -0.06 | 0.16 | 0.07 | 1.6 | 0.25 | 0.30 |
| AI | -0.23 | -0.13 | -0.10 | 0.13 | -0.10 | 1.03 | -0.02 | 0.33 | 0.18 | 1.5 | 0.13 | 0.10 |
| PI | 0.26 | 0.02 | 0.13 | -0.12 | -0.20 | 0.09 | -0.34 | 0.80 | -0.06 | 1.9 | 0.25 | 0.11 |
| C1a | -0.29 | 0.00 | -0.08 | -0.23 | -0.03 | -0.31 | 0.09 | -0.80 | -0.01 | 1.8 | 0.24 | 0.25 |
| C2a | 0.32 | 0.09 | 0.08 | 0.06 | -0.07 | 0.23 | 0.04 | 0.93 | -0.09 | 1.5 | 0.13 | 0.17 |
| Ca | 0.26 | 0.12 | 0.16 | -0.09 | -0.14 | 0.17 | -0.02 | 0.92 | -0.12 | 1.4 | 0.19 | 0.14 |
| DFA_alpha1 | -0.22 | 0.10 | 1.02 | -0.15 | 0.07 | -0.09 | 0.04 | 0.24 | 0.05 | 1.3 | 0.05 | 0.17 |
| DFA_alpha1_ExpRange | 0.04 | 0.15 | -0.01 | -0.21 | 0.13 | 0.19 | 0.04 | -0.04 | 0.83 | 1.4 | 0.10 | 0.16 |
| DFA_alpha1_ExpMean | -0.20 | 0.59 | 0.32 | -0.20 | -0.09 | 0.08 | 0.15 | 0.10 | 0.30 | 3.1 | 0.20 | 0.19 |
| DFA_alpha1_DimRange | 0.07 | 0.15 | -0.11 | -0.23 | 0.13 | 0.19 | 0.14 | -0.30 | 0.75 | 2.0 | 0.08 | 0.75 |
| DFA_alpha1_DimMean | -0.22 | 0.64 | -0.10 | 0.18 | -0.34 | -0.10 | 0.05 | 0.11 | -0.79 | 2.8 | 0.23 | 0.10 |
| DFA_alpha2 | 0.23 | 0.05 | 0.01 | 0.37 | -0.60 | 0.21 | 0.57 | -0.13 | 0.05 | 3.4 | 0.18 | 0.12 |
| DFA_alpha2_ExpRange | -0.23 | -0.24 | -0.05 | -0.12 | -0.12 | -0.78 | -0.20 | -0.03 | -0.10 | 1.7 | 0.30 | 0.08 |
| DFA_alpha2_ExpMean | 0.15 | -0.15 | 0.15 | 0.39 | -0.45 | -0.03 | 0.56 | -0.13 | -0.12 | 3.6 | 0.16 | 0.58 |
| DFA_alpha2_DimRange | 0.03 | -0.01 | 0.01 | 0.01 | -0.30 | -0.66 | 0.06 | 0.02 | 0.11 | 1.5 | 0.39 | 0.08 |
| DFA_alpha2_DimMean | 0.06 | 0.65 | 0.06 | 0.24 | -0.25 | -0.19 | -0.20 | -0.02 | 0.34 | 2.8 | 0.31 | 0.11 |
| ApEn | 0.14 | 0.60 | 0.13 | -0.19 | 0.31 | 0.28 | -0.23 | 0.06 | -0.06 | 3.0 | 0.33 | 0.23 |
| SampEn | -0.12 | 0.84 | 0.01 | 0.12 | 0.20 | -0.12 | 0.11 | 0.06 | -0.06 | 1.3 | 0.16 | 0.27 |
| ShanEn | 0.14 | 0.13 | -0.15 | -0.37 | 0.20 | 0.18 | 0.53 | -0.16 | 0.05 | 3.2 | 0.14 | 0.45 |
| FuzzyEn | -0.05 | 0.81 | -0.16 | -0.08 | 0.11 | -0.12 | -0.04 | -0.01 | 0.04 | 1.2 | 0.08 | 0.29 |
| MSE | 0.20 | -0.18 | 0.18 | -0.28 | 0.40 | 0.26 | 0.25 | -0.14 | -0.22 | 6.2 | 0.25 | 0.25 |
| CMSE | 0.11 | -0.08 | 0.19 | -0.07 | 0.00 | -0.02 | 0.87 | -0.19 | -0.08 | 1.3 | 0.19 | 0.12 |
| RCMSE | 0.12 | 0.02 | 0.15 | 0.01 | -0.17 | -0.03 | 0.81 | -0.17 | 0.12 | 1.4 | 0.24 | 0.50 |
| CD | -0.14 | 0.97 | 0.23 | 0.05 | -0.08 | 0.03 | 0.12 | 0.07 | -0.06 | 1.2 | 0.07 | 0.19 |
| HFD | -0.03 | 0.58 | -0.55 | -0.01 | 0.00 | -0.03 | -0.27 | -0.25 | 0.03 | 2.8 | 0.13 | 0.20 |
| KFD | 0.09 | 0.99 | 0.12 | -0.02 | 0.05 | 0.05 | -0.22 | -0.01 | -0.11 | 1.2 | 0.22 | 0.15 |
| LZC | -0.09 | 0.59 | -0.14 | 0.07 | 0.06 | -0.12 | 0.35 | 0.28 | -0.06 | 2.5 | 0.14 | 0.37 |
# insight::display(pca, threshold="max", sort=TRUE)
plot(pca, size_text = 3) +
theme(axis.text.y=element_text(size=5))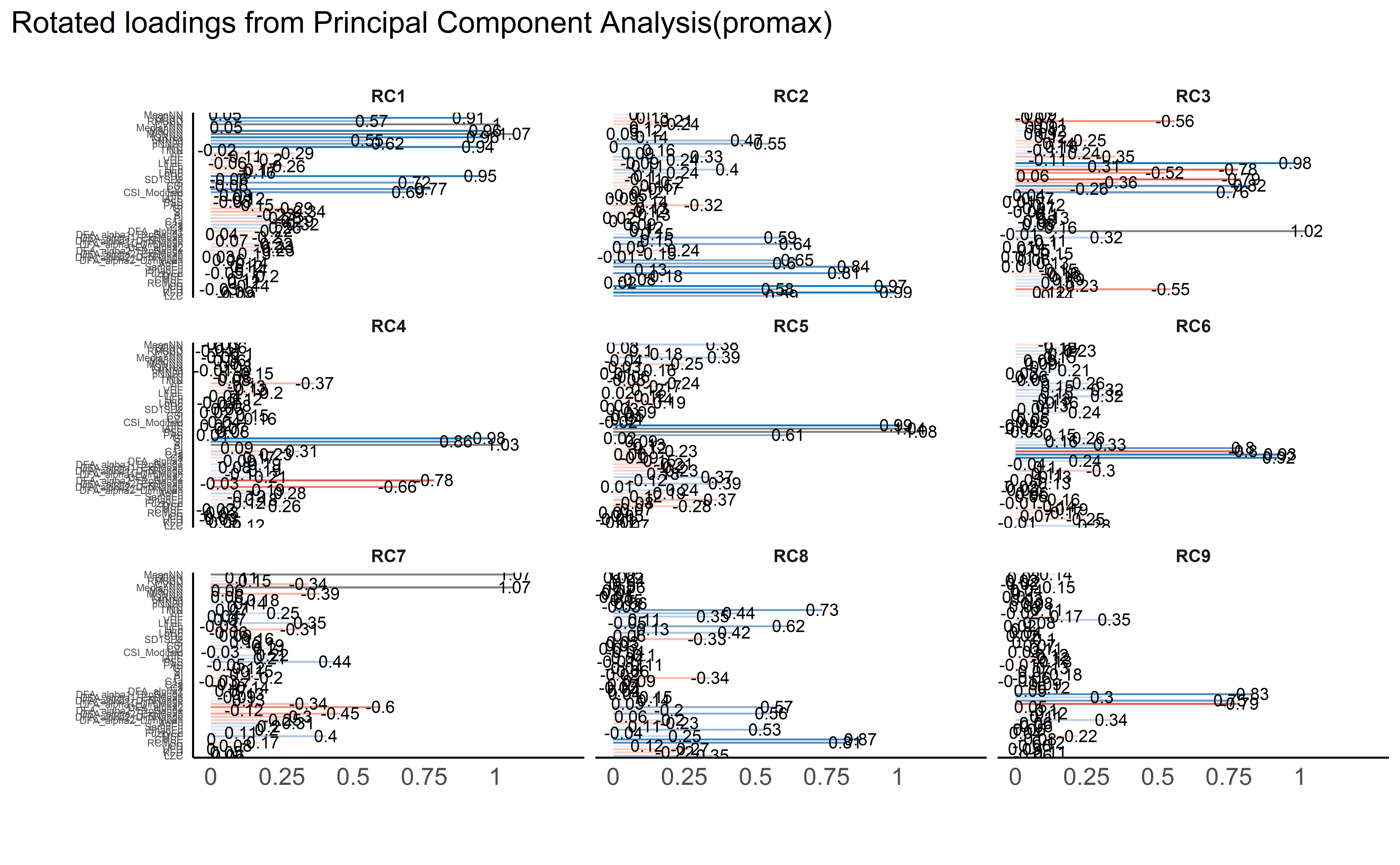
Figure 1: PCA 9-component Rotated Loadings
table_clusters <- parameters::closest_component(pca) %>%
data.frame(Index = names(.),
PCA_9 = .) %>%
mutate(PCA_9 = as.character(PCA_9)) %>%
datawizard::data_rename_rows()12-components
set.seed(3)
pca <- parameters::principal_components(data[hrv_cols], n=12, rotation = "promax")
knitr::kable(pca,
booktabs = TRUE,
caption = "Item loadings for PCA 12-component solutions."
)| Variable | RC1 | RC2 | RC3 | RC5 | RC8 | RC6 | RC7 | RC4 | RC10 | RC12 | RC11 | RC9 | Complexity | Uniqueness | MSA |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MeanNN | 0.02 | 0.01 | 0.09 | 0.28 | 0.07 | -0.01 | 1.07 | -0.10 | -0.03 | -0.07 | 0.05 | 0.09 | 1.2 | 0.04 | 0.17 |
| SDNN | 0.93 | -0.11 | -0.02 | 0.05 | -0.04 | 0.04 | 0.07 | 0.00 | 0.00 | 0.04 | -0.09 | 0.00 | 1.1 | 0.01 | 0.28 |
| RMSSD | 0.61 | -0.09 | -0.46 | 0.15 | -0.18 | 0.09 | 0.00 | -0.01 | 0.00 | 0.16 | -0.21 | 0.03 | 2.8 | 0.03 | 0.29 |
| CVNN | 1.03 | -0.25 | -0.07 | -0.13 | -0.08 | 0.02 | -0.35 | 0.03 | -0.04 | 0.10 | -0.09 | -0.06 | 1.5 | 0.02 | 0.27 |
| MedianNN | 0.02 | -0.01 | 0.10 | 0.28 | 0.07 | -0.01 | 1.07 | -0.11 | -0.04 | -0.05 | 0.07 | 0.09 | 1.2 | 0.04 | 0.15 |
| MadNN | 0.93 | 0.14 | 0.12 | -0.04 | 0.10 | 0.05 | 0.11 | 0.06 | 0.02 | -0.20 | 0.08 | 0.04 | 1.3 | 0.02 | 0.24 |
| MCVNN | 1.04 | 0.05 | 0.07 | -0.23 | 0.08 | 0.02 | -0.29 | 0.11 | -0.01 | -0.19 | 0.09 | 0.00 | 1.4 | 0.05 | 0.31 |
| IQRNN | 0.92 | 0.13 | 0.12 | -0.04 | 0.08 | 0.08 | 0.11 | 0.05 | 0.00 | -0.18 | 0.08 | 0.01 | 1.2 | 0.03 | 0.24 |
| pNN50 | 0.60 | 0.58 | -0.19 | 0.19 | 0.02 | 0.09 | 0.15 | 0.10 | 0.10 | -0.07 | 0.02 | -0.04 | 2.8 | 0.10 | 0.25 |
| pNN20 | 0.67 | 0.55 | -0.09 | 0.04 | -0.02 | -0.08 | 0.08 | 0.05 | -0.05 | 0.11 | 0.08 | -0.02 | 2.2 | 0.06 | 0.25 |
| HTI | 0.89 | 0.04 | 0.16 | -0.04 | 0.02 | 0.02 | 0.06 | -0.03 | -0.06 | -0.15 | 0.00 | 0.07 | 1.2 | 0.04 | 0.26 |
| TINN | 0.10 | 0.03 | -0.06 | -0.13 | 0.54 | -0.10 | 0.28 | 0.02 | 0.17 | 0.31 | -0.03 | -0.11 | 2.9 | 0.14 | 0.47 |
| LF | -0.17 | 0.21 | 0.35 | -0.19 | 0.08 | 0.08 | 0.09 | 0.14 | -0.19 | 0.46 | -0.36 | -0.06 | 5.1 | 0.10 | 0.18 |
| HF | -0.18 | 0.29 | -0.37 | -0.27 | 0.35 | 0.27 | 0.22 | 0.06 | -0.10 | -0.06 | 0.03 | -0.16 | 6.8 | 0.23 | 0.18 |
| VHF | -0.16 | 0.07 | -0.04 | 0.13 | -0.22 | 0.18 | -0.08 | -0.08 | -0.23 | 0.98 | 0.08 | 0.09 | 1.5 | 0.16 | 0.46 |
| LFHF | 0.05 | -0.06 | 1.03 | 0.07 | -0.21 | -0.02 | 0.18 | 0.08 | -0.06 | 0.23 | -0.16 | 0.05 | 1.4 | 0.16 | 0.13 |
| LFn | 0.02 | 0.14 | 0.47 | -0.02 | 0.23 | -0.02 | -0.15 | 0.24 | 0.22 | 0.42 | -0.46 | -0.05 | 4.9 | 0.09 | 0.14 |
| HFn | -0.21 | 0.28 | -0.81 | -0.21 | 0.02 | 0.01 | -0.10 | 0.05 | 0.09 | -0.15 | 0.30 | -0.06 | 2.0 | 0.05 | 0.22 |
| LnHF | -0.09 | 0.36 | -0.45 | -0.18 | 0.25 | 0.09 | -0.06 | 0.11 | 0.06 | 0.22 | -0.13 | -0.01 | 4.2 | 0.06 | 0.25 |
| SD2 | 0.96 | -0.10 | 0.10 | 0.02 | 0.02 | 0.03 | 0.08 | 0.02 | 0.00 | 0.00 | -0.06 | -0.01 | 1.1 | 0.01 | 0.27 |
| SD1SD2 | -0.03 | -0.12 | -0.71 | 0.05 | -0.44 | 0.02 | 0.04 | 0.00 | -0.07 | 0.26 | -0.05 | 0.05 | 2.1 | 0.05 | 0.20 |
| S | 0.75 | 0.03 | -0.28 | 0.18 | -0.06 | 0.09 | -0.06 | 0.01 | 0.05 | 0.00 | -0.26 | 0.08 | 1.8 | 0.06 | 0.66 |
| CSI | -0.11 | 0.06 | 0.73 | 0.04 | 0.15 | 0.04 | -0.09 | -0.08 | 0.09 | -0.16 | 0.15 | -0.07 | 1.5 | 0.10 | 0.49 |
| CVI | 0.81 | -0.26 | -0.19 | -0.06 | -0.11 | -0.02 | 0.20 | 0.02 | -0.09 | 0.20 | -0.05 | -0.01 | 1.7 | 0.03 | 0.48 |
| CSI_Modified | 0.64 | -0.13 | 0.69 | -0.08 | 0.05 | -0.03 | 0.12 | -0.09 | -0.04 | -0.06 | 0.24 | -0.08 | 2.5 | 0.05 | 0.59 |
| PIP | -0.06 | -0.03 | -0.03 | 1.01 | -0.02 | -0.03 | 0.15 | 0.01 | -0.02 | 0.04 | 0.10 | -0.08 | 1.1 | 0.03 | 0.26 |
| IALS | -0.09 | 0.08 | -0.05 | 1.07 | 0.01 | -0.02 | 0.12 | 0.01 | 0.03 | 0.07 | 0.08 | -0.09 | 1.1 | 0.04 | 0.28 |
| PSS | 0.06 | 0.24 | 0.07 | 1.13 | -0.02 | -0.16 | 0.30 | 0.04 | 0.13 | 0.03 | -0.01 | -0.11 | 1.4 | 0.07 | 0.11 |
| PAS | -0.23 | -0.28 | 0.08 | 0.61 | 0.01 | 0.20 | -0.08 | 0.04 | -0.09 | 0.08 | 0.11 | 0.07 | 2.3 | 0.16 | 0.19 |
| GI | -0.03 | -0.03 | 0.02 | 0.06 | -0.07 | 0.03 | -0.02 | -0.07 | 0.99 | -0.14 | -0.14 | 0.03 | 1.1 | 0.02 | 0.15 |
| SI | -0.03 | -0.01 | 0.06 | -0.05 | -0.08 | -0.07 | -0.01 | 0.06 | 0.99 | -0.25 | -0.16 | -0.03 | 1.2 | 0.03 | 0.30 |
| AI | -0.05 | -0.06 | -0.04 | 0.16 | -0.05 | 0.15 | -0.04 | -0.20 | 0.91 | -0.03 | -0.11 | 0.09 | 1.3 | 0.07 | 0.10 |
| PI | 0.11 | -0.03 | 0.07 | -0.21 | -0.14 | 0.87 | 0.03 | 0.17 | 0.08 | -0.14 | 0.25 | 0.01 | 1.6 | 0.16 | 0.11 |
| C1a | -0.09 | 0.01 | -0.01 | -0.18 | 0.01 | -0.85 | -0.04 | 0.19 | -0.05 | -0.12 | -0.10 | -0.08 | 1.3 | 0.17 | 0.25 |
| C2a | 0.11 | 0.04 | 0.02 | 0.00 | 0.04 | 0.96 | -0.04 | -0.22 | -0.03 | 0.23 | 0.04 | -0.04 | 1.3 | 0.05 | 0.17 |
| Ca | 0.10 | 0.06 | 0.11 | -0.16 | 0.01 | 0.93 | -0.01 | -0.06 | 0.03 | 0.12 | 0.07 | -0.08 | 1.2 | 0.14 | 0.14 |
| DFA_alpha1 | -0.14 | 0.13 | 1.03 | -0.12 | -0.05 | 0.12 | 0.03 | 0.13 | 0.04 | 0.12 | -0.05 | 0.03 | 1.2 | 0.04 | 0.17 |
| DFA_alpha1_ExpRange | 0.00 | 0.18 | 0.00 | -0.18 | -0.02 | -0.05 | -0.01 | -0.05 | 0.06 | 0.48 | 0.25 | 0.67 | 2.6 | 0.08 | 0.16 |
| DFA_alpha1_ExpMean | -0.08 | 0.44 | 0.36 | -0.18 | -0.07 | -0.05 | -0.13 | -0.05 | 0.08 | 0.55 | 0.14 | 0.05 | 3.5 | 0.14 | 0.19 |
| DFA_alpha1_DimRange | 0.01 | 0.19 | -0.13 | -0.22 | 0.17 | -0.22 | 0.07 | -0.06 | 0.07 | 0.23 | 0.26 | 0.63 | 2.9 | 0.07 | 0.75 |
| DFA_alpha1_DimMean | -0.06 | 0.33 | -0.07 | 0.14 | -0.14 | -0.04 | -0.19 | -0.10 | -0.01 | 0.10 | -0.08 | -0.92 | 1.6 | 0.10 | 0.10 |
| DFA_alpha2 | 0.11 | -0.07 | -0.10 | 0.32 | 0.69 | -0.02 | -0.40 | -0.16 | -0.02 | 0.13 | 0.27 | 0.00 | 2.9 | 0.14 | 0.12 |
| DFA_alpha2_ExpRange | -0.02 | 0.03 | 0.07 | -0.06 | -0.20 | -0.17 | -0.08 | 0.87 | -0.18 | -0.15 | -0.07 | -0.02 | 1.4 | 0.17 | 0.08 |
| DFA_alpha2_ExpMean | 0.09 | -0.01 | 0.09 | 0.44 | 0.60 | -0.11 | -0.47 | -0.03 | -0.13 | 0.01 | -0.04 | -0.01 | 3.1 | 0.11 | 0.58 |
| DFA_alpha2_DimRange | 0.21 | 0.21 | 0.10 | 0.04 | 0.14 | -0.10 | -0.11 | 0.94 | -0.06 | -0.08 | 0.19 | 0.10 | 1.5 | 0.22 | 0.08 |
| DFA_alpha2_DimMean | -0.10 | 0.36 | -0.05 | 0.12 | 0.10 | 0.20 | 0.06 | 0.25 | -0.19 | 0.11 | 0.83 | 0.18 | 2.2 | 0.22 | 0.11 |
| ApEn | 0.02 | 0.88 | 0.10 | -0.02 | -0.27 | 0.02 | -0.18 | -0.51 | -0.03 | -0.21 | -0.26 | 0.17 | 2.4 | 0.06 | 0.23 |
| SampEn | -0.09 | 1.12 | 0.03 | 0.21 | 0.10 | -0.01 | 0.02 | 0.11 | -0.03 | -0.15 | 0.00 | 0.05 | 1.2 | 0.04 | 0.27 |
| ShanEn | 0.10 | 0.07 | -0.16 | -0.38 | 0.33 | -0.12 | 0.12 | -0.42 | -0.03 | 0.17 | -0.24 | -0.02 | 4.9 | 0.12 | 0.45 |
| FuzzyEn | -0.01 | 0.92 | -0.13 | -0.03 | -0.04 | -0.05 | 0.04 | 0.14 | -0.01 | -0.07 | 0.13 | 0.03 | 1.2 | 0.04 | 0.29 |
| MSE | -0.02 | -0.07 | 0.08 | -0.25 | 0.20 | 0.00 | 0.12 | -0.70 | -0.14 | -0.28 | -0.45 | 0.01 | 2.9 | 0.13 | 0.25 |
| CMSE | -0.02 | 0.04 | 0.10 | -0.11 | 0.89 | -0.03 | 0.05 | -0.14 | -0.13 | -0.18 | -0.16 | 0.05 | 1.3 | 0.14 | 0.12 |
| RCMSE | -0.02 | 0.07 | 0.04 | -0.09 | 0.98 | 0.04 | 0.09 | 0.11 | -0.04 | -0.18 | 0.18 | 0.15 | 1.3 | 0.15 | 0.50 |
| CD | -0.14 | 0.82 | 0.19 | 0.02 | 0.11 | 0.07 | 0.02 | -0.05 | 0.02 | 0.10 | 0.29 | -0.18 | 1.7 | 0.06 | 0.19 |
| HFD | 0.01 | 0.41 | -0.53 | -0.03 | -0.26 | -0.23 | 0.05 | -0.01 | -0.01 | 0.03 | 0.25 | -0.10 | 3.5 | 0.12 | 0.20 |
| KFD | 0.07 | 0.75 | 0.08 | -0.02 | -0.28 | -0.03 | -0.01 | -0.31 | -0.09 | 0.15 | 0.21 | -0.24 | 2.3 | 0.19 | 0.15 |
| LZC | -0.01 | 0.84 | -0.06 | 0.16 | 0.17 | 0.12 | -0.10 | 0.14 | -0.01 | 0.17 | -0.15 | -0.02 | 1.5 | 0.08 | 0.37 |
# insight::display(pca, threshold="max", sort=TRUE)
plot(pca, size_text = 3) +
theme(axis.text.y=element_text(size=5))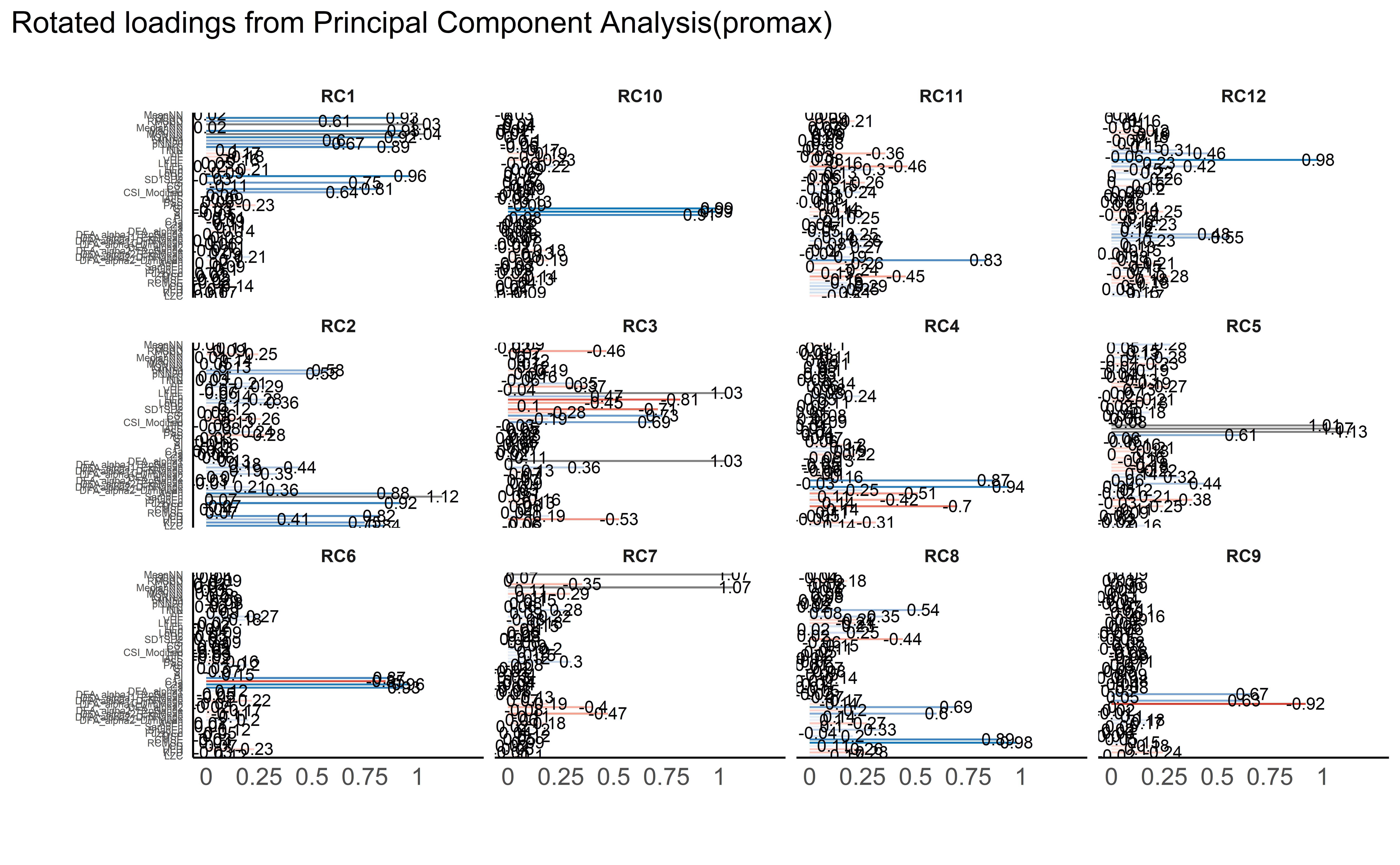
Figure 2: PCA 12-component Rotated Loadings
table_clusters$PCA_12 <- as.character(parameters::closest_component(pca))Factor Analysis
How many factors
set.seed(3)
# n_max to 28 to have at least 2 var per component
n <- parameters::n_factors(data[hrv_cols], cor=r, type = "FA", package = "all", n_max = 28)
n> # Method Agreement Procedure:
>
> The choice of 1 dimensions is supported by 4 (25.00%) methods out of 16 (t, p, TLI, RMSEA).plot(n)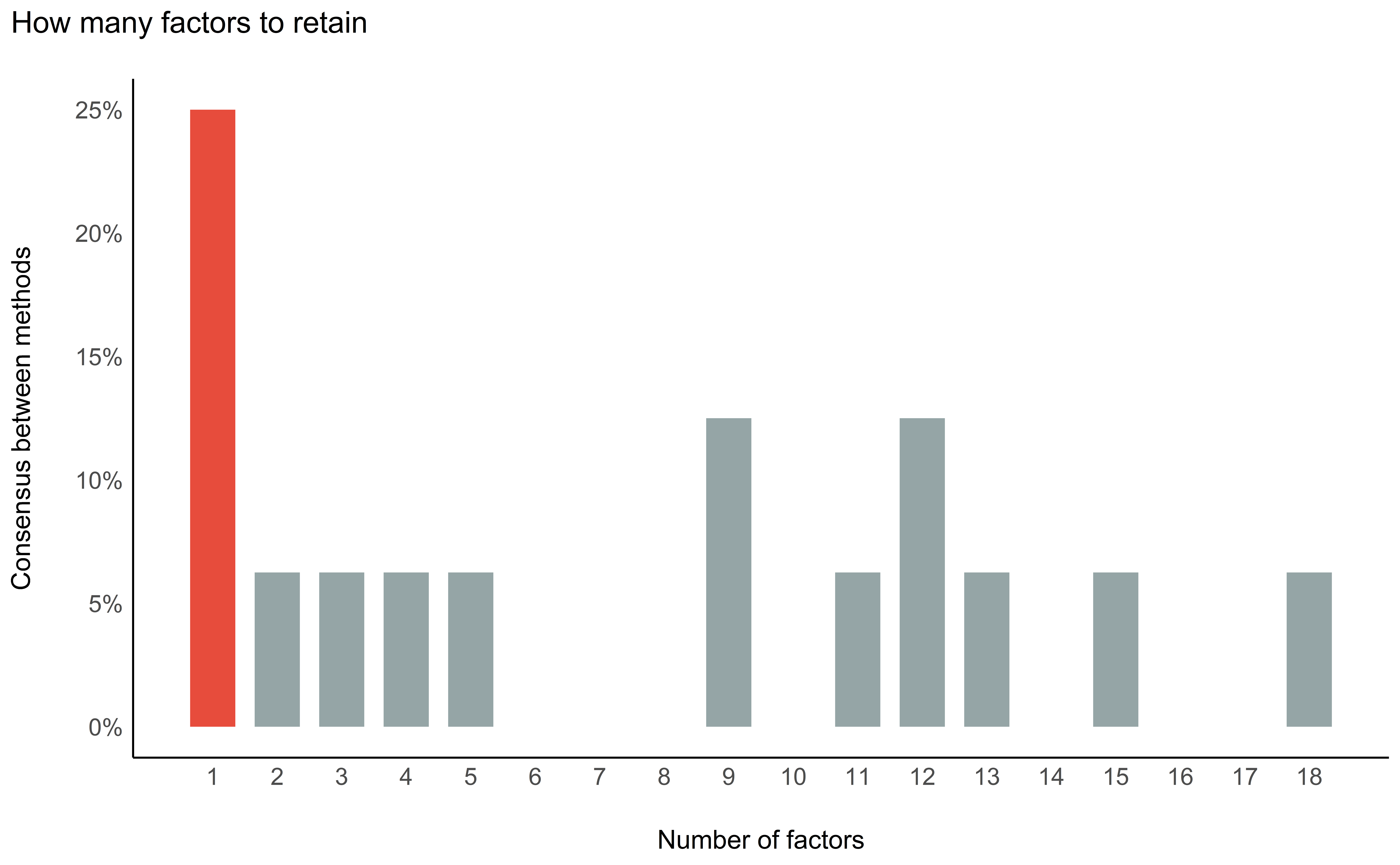
9-factors
set.seed(3)
efa <- parameters::factor_analysis(data[hrv_cols], cor=r, n=9, rotation="promax", fm="ml")
knitr::kable(efa,
booktabs = TRUE,
caption = "Item loadings for EFA 9-factor solutions."
)| Variable | ML1 | ML5 | ML4 | ML2 | ML8 | ML3 | ML7 | ML9 | ML6 | Complexity | Uniqueness |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MeanNN | -0.11 | -0.10 | -0.10 | 0.11 | 0.07 | 1.11 | -0.04 | -0.10 | -0.05 | 1.1 | 0.00 |
| SDNN | 0.88 | 0.14 | -0.05 | 0.05 | -0.02 | -0.05 | -0.09 | 0.00 | 0.50 | 1.7 | 0.00 |
| RMSSD | 0.40 | -0.33 | -0.19 | -0.03 | 0.06 | 0.23 | 0.05 | 0.08 | 0.37 | 4.3 | 0.15 |
| CVNN | 0.78 | 0.25 | -0.02 | 0.05 | -0.14 | -0.31 | 0.08 | -0.24 | 0.31 | 2.3 | 0.10 |
| MedianNN | -0.09 | -0.01 | -0.02 | 0.08 | 0.02 | 1.04 | -0.03 | 0.01 | 0.02 | 1.0 | 0.02 |
| MadNN | 1.04 | 0.05 | 0.02 | -0.01 | 0.09 | 0.01 | 0.00 | 0.12 | -0.21 | 1.1 | 0.00 |
| MCVNN | 1.09 | 0.10 | -0.01 | -0.10 | 0.06 | -0.38 | 0.08 | -0.03 | -0.29 | 1.5 | 0.06 |
| IQRNN | 1.07 | 0.10 | 0.05 | -0.03 | 0.05 | -0.07 | 0.01 | 0.13 | -0.15 | 1.1 | 0.02 |
| pNN50 | 0.51 | -0.19 | 0.34 | 0.18 | 0.07 | 0.29 | 0.02 | -0.13 | 0.03 | 3.4 | 0.21 |
| pNN20 | 0.51 | -0.28 | 0.44 | 0.09 | 0.01 | 0.21 | 0.01 | 0.06 | -0.04 | 3.0 | 0.09 |
| HTI | 0.88 | 0.02 | 0.02 | -0.03 | -0.08 | 0.05 | 0.01 | 0.24 | -0.06 | 1.2 | 0.13 |
| TINN | -0.14 | 0.13 | 0.51 | -0.25 | 0.18 | 0.12 | -0.06 | -0.24 | -0.04 | 2.9 | 0.45 |
| LF | 0.02 | 0.03 | 0.27 | -0.17 | 0.67 | -0.02 | -0.07 | 0.05 | 0.19 | 1.7 | 0.34 |
| HF | 0.02 | -0.42 | 0.40 | -0.10 | 0.31 | -0.16 | 0.00 | -0.04 | 0.06 | 3.4 | 0.45 |
| VHF | -0.14 | -0.42 | 0.32 | 0.07 | 0.40 | -0.08 | 0.05 | 0.01 | -0.10 | 3.4 | 0.52 |
| LFHF | 0.09 | 0.71 | -0.03 | 0.03 | 0.32 | -0.03 | 0.01 | 0.00 | 0.07 | 1.5 | 0.36 |
| LFn | -0.02 | 0.67 | 0.17 | -0.06 | 0.47 | -0.02 | -0.01 | -0.10 | 0.10 | 2.1 | 0.21 |
| HFn | -0.17 | -0.72 | 0.23 | -0.12 | -0.22 | -0.09 | 0.01 | -0.11 | -0.10 | 1.7 | 0.36 |
| LnHF | -0.11 | -0.60 | 0.33 | -0.24 | 0.32 | -0.07 | -0.04 | 0.00 | 0.13 | 2.8 | 0.22 |
| SD2 | 0.92 | 0.17 | -0.09 | 0.01 | 0.02 | -0.05 | -0.06 | -0.02 | 0.30 | 1.3 | 0.11 |
| SD1SD2 | -0.03 | -1.05 | -0.38 | 0.00 | 0.15 | -0.02 | -0.04 | 0.09 | -0.01 | 1.3 | 0.10 |
| S | 0.61 | -0.09 | -0.06 | -0.11 | 0.04 | 0.09 | 0.00 | 0.13 | 0.47 | 2.2 | 0.16 |
| CSI | 0.09 | 0.84 | 0.13 | -0.13 | -0.12 | -0.10 | -0.05 | 0.07 | -0.12 | 1.3 | 0.20 |
| CVI | 0.57 | -0.35 | -0.23 | 0.01 | 0.09 | 0.21 | -0.07 | -0.21 | 0.23 | 3.3 | 0.08 |
| CSI_Modified | 0.40 | 0.39 | -0.27 | -0.11 | 0.05 | 0.26 | 0.10 | -0.30 | -0.38 | 5.7 | 0.38 |
| PIP | -0.04 | 0.03 | -0.06 | 1.02 | 0.03 | 0.05 | -0.06 | 0.07 | -0.01 | 1.0 | 0.00 |
| IALS | -0.04 | 0.05 | -0.01 | 1.01 | 0.01 | 0.05 | -0.06 | 0.00 | 0.00 | 1.0 | 0.02 |
| PSS | -0.04 | 0.12 | 0.07 | 1.05 | 0.06 | 0.22 | -0.04 | 0.09 | 0.02 | 1.2 | 0.13 |
| PAS | 0.01 | 0.10 | -0.20 | 0.69 | 0.02 | -0.17 | 0.00 | -0.08 | 0.04 | 1.4 | 0.19 |
| GI | -0.12 | 0.04 | -0.11 | -0.21 | -0.47 | -0.12 | -0.07 | -0.03 | 0.02 | 1.9 | 0.77 |
| SI | -0.04 | 0.14 | -0.11 | -0.16 | -0.37 | 0.04 | -0.33 | 0.02 | 0.00 | 3.0 | 0.69 |
| AI | -0.08 | -0.15 | -0.04 | 0.02 | -0.44 | -0.03 | -0.08 | -0.15 | 0.03 | 1.7 | 0.75 |
| PI | 0.11 | 0.08 | -0.05 | -0.02 | 0.04 | -0.10 | 0.70 | -0.14 | 0.00 | 1.2 | 0.37 |
| C1a | 0.02 | 0.26 | 0.02 | 0.18 | 0.06 | 0.01 | -0.58 | -0.09 | -0.04 | 1.7 | 0.56 |
| C2a | -0.04 | 0.00 | 0.06 | -0.01 | 0.02 | -0.06 | 0.99 | 0.05 | -0.05 | 1.0 | 0.08 |
| Ca | -0.08 | 0.18 | 0.03 | -0.02 | 0.10 | 0.08 | 0.90 | 0.00 | -0.09 | 1.2 | 0.28 |
| DFA_alpha1 | -0.05 | 0.87 | 0.21 | -0.19 | 0.20 | 0.03 | 0.08 | 0.03 | -0.01 | 1.4 | 0.10 |
| DFA_alpha1_ExpRange | 0.07 | -0.08 | 0.50 | -0.05 | -0.28 | 0.06 | 0.20 | 0.07 | 0.06 | 2.2 | 0.55 |
| DFA_alpha1_ExpMean | -0.16 | 0.42 | 0.80 | -0.10 | -0.10 | 0.10 | 0.15 | -0.06 | -0.01 | 1.8 | 0.34 |
| DFA_alpha1_DimRange | -0.21 | 0.11 | -0.01 | 0.06 | -0.28 | 0.03 | 0.13 | 0.04 | 0.04 | 3.0 | 0.88 |
| DFA_alpha1_DimMean | -0.10 | 0.33 | 0.20 | 0.11 | 0.27 | -0.18 | -0.10 | -0.34 | -0.05 | 4.9 | 0.43 |
| DFA_alpha2 | -0.05 | 0.64 | 0.15 | 0.23 | -0.11 | -0.01 | -0.05 | -0.10 | -0.08 | 1.6 | 0.52 |
| DFA_alpha2_ExpRange | -0.17 | 0.06 | -0.17 | -0.05 | 0.65 | 0.05 | 0.01 | -0.16 | -0.03 | 1.5 | 0.46 |
| DFA_alpha2_ExpMean | -0.01 | 0.49 | 0.00 | 0.07 | 0.11 | 0.00 | -0.01 | -0.02 | -0.06 | 1.2 | 0.69 |
| DFA_alpha2_DimRange | -0.06 | 0.15 | -0.04 | -0.02 | 0.57 | -0.08 | -0.05 | -0.12 | -0.03 | 1.4 | 0.51 |
| DFA_alpha2_DimMean | 0.01 | -0.01 | 0.31 | 0.11 | -0.30 | -0.08 | -0.01 | 0.04 | 0.00 | 2.4 | 0.84 |
| ApEn | 0.16 | -0.08 | 0.50 | 0.03 | -0.46 | -0.08 | 0.01 | 0.66 | 0.13 | 3.1 | 0.14 |
| SampEn | 0.05 | 0.00 | 0.80 | 0.11 | 0.05 | -0.09 | -0.07 | 0.70 | 0.13 | 2.1 | 0.02 |
| ShanEn | 0.25 | -0.14 | 0.31 | -0.27 | -0.39 | 0.20 | -0.03 | -0.24 | -0.10 | 5.6 | 0.30 |
| FuzzyEn | 0.07 | -0.18 | 0.83 | 0.05 | 0.02 | -0.10 | -0.06 | 0.46 | 0.06 | 1.8 | 0.05 |
| MSE | 0.07 | -0.02 | 0.05 | -0.07 | -0.59 | 0.20 | -0.10 | 0.27 | 0.03 | 1.8 | 0.37 |
| CMSE | -0.07 | 0.28 | 0.41 | -0.23 | 0.08 | 0.12 | -0.10 | 0.34 | -0.02 | 4.0 | 0.39 |
| RCMSE | -0.07 | 0.36 | 0.44 | -0.22 | 0.21 | 0.12 | -0.09 | 0.26 | -0.03 | 4.1 | 0.35 |
| CD | 0.05 | 0.26 | 0.94 | 0.02 | 0.06 | -0.15 | -0.01 | 0.15 | -0.11 | 1.3 | 0.16 |
| HFD | -0.13 | -0.80 | 0.16 | 0.06 | -0.05 | 0.11 | -0.08 | -0.04 | -0.08 | 1.2 | 0.25 |
| KFD | 0.11 | -0.36 | 0.55 | 0.09 | -0.28 | -0.03 | -0.11 | 0.02 | -0.02 | 2.5 | 0.32 |
| LZC | -0.05 | -0.29 | 0.63 | 0.01 | 0.26 | 0.10 | 0.00 | 0.17 | 0.16 | 2.2 | 0.21 |
# insight::display(efa, threshold="max", sort=TRUE)
plot(efa, size_text = 3) +
theme(axis.text.y=element_text(size=5))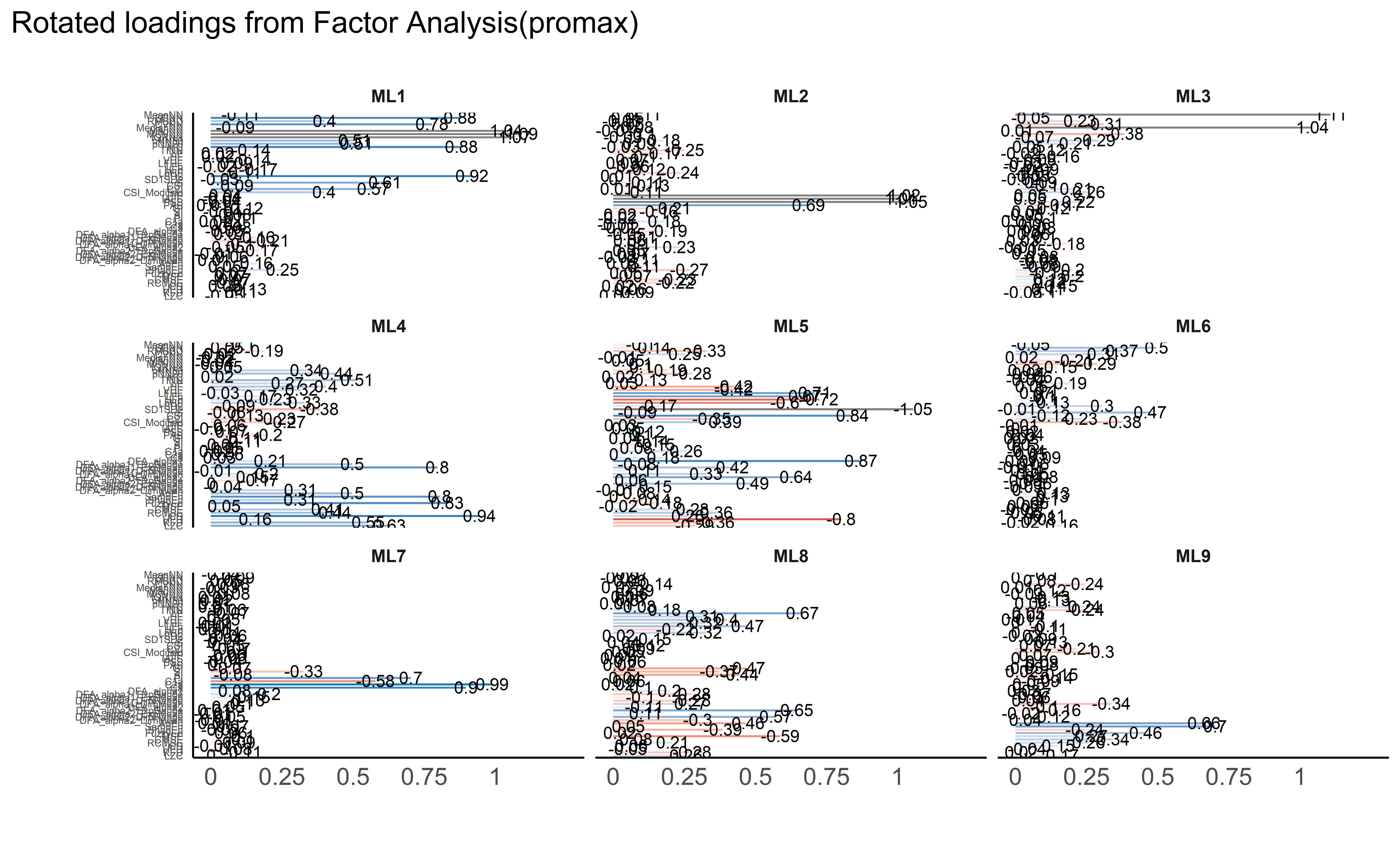
Figure 3: EFA 9-factor Rotated Loadings
table_clusters$EFA_9 <- as.character(parameters::closest_component(efa))12-factors
set.seed(3)
efa <- parameters::factor_analysis(data[hrv_cols], cor=r, n=12, rotation="promax", fm="ml")
knitr::kable(efa,
booktabs = TRUE,
caption = "Item loadings for EFA 12-factor solutions."
)| Variable | ML1 | ML4 | ML5 | ML8 | ML2 | ML3 | ML7 | ML9 | ML12 | ML10 | ML11 | ML6 | Complexity | Uniqueness |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MeanNN | -0.03 | -0.05 | -0.13 | 0.06 | 0.14 | 1.06 | -0.02 | -0.02 | 0.04 | -0.01 | 0.06 | 0.08 | 1.1 | 0.01 |
| SDNN | 0.82 | 0.16 | -0.10 | -0.02 | 0.04 | -0.03 | -0.08 | -0.02 | 0.06 | 0.25 | 0.44 | -0.16 | 2.0 | 0.00 |
| RMSSD | 0.36 | -0.29 | -0.13 | -0.09 | 0.01 | 0.15 | 0.10 | -0.01 | 0.13 | -0.03 | 0.43 | -0.05 | 3.9 | 0.14 |
| CVNN | 0.68 | 0.31 | -0.18 | -0.03 | -0.01 | -0.21 | 0.03 | -0.06 | -0.04 | 0.57 | 0.26 | -0.11 | 3.3 | 0.06 |
| MedianNN | 0.00 | 0.05 | -0.09 | 0.01 | 0.11 | 1.06 | -0.03 | -0.06 | 0.04 | 0.01 | 0.07 | -0.05 | 1.1 | 0.02 |
| MadNN | 1.03 | 0.01 | 0.02 | 0.03 | 0.04 | 0.11 | -0.02 | 0.01 | 0.11 | -0.07 | -0.27 | 0.08 | 1.2 | 0.00 |
| MCVNN | 1.04 | 0.05 | -0.05 | 0.06 | -0.08 | -0.26 | 0.05 | 0.01 | 0.11 | 0.09 | -0.34 | 0.13 | 1.5 | 0.06 |
| IQRNN | 1.07 | 0.05 | 0.03 | 0.03 | -0.01 | 0.06 | -0.02 | -0.01 | 0.06 | -0.04 | -0.25 | 0.02 | 1.1 | 0.02 |
| pNN50 | 0.50 | -0.17 | 0.35 | 0.05 | 0.20 | 0.26 | 0.01 | 0.01 | -0.03 | 0.15 | 0.05 | 0.08 | 3.5 | 0.22 |
| pNN20 | 0.45 | -0.27 | 0.51 | -0.11 | 0.08 | 0.14 | -0.02 | -0.09 | -0.06 | -0.01 | 0.00 | 0.17 | 3.3 | 0.05 |
| HTI | 0.84 | 0.00 | 0.06 | -0.20 | 0.01 | 0.06 | 0.02 | 0.01 | 0.07 | -0.18 | -0.09 | 0.08 | 1.3 | 0.12 |
| TINN | -0.15 | 0.12 | 0.50 | 0.26 | -0.19 | 0.05 | -0.01 | 0.10 | 0.18 | 0.19 | 0.05 | 0.13 | 3.3 | 0.42 |
| LF | 0.12 | -0.09 | 0.45 | 0.68 | -0.09 | -0.02 | -0.06 | -0.01 | 0.06 | -0.18 | 0.12 | -0.09 | 2.2 | 0.30 |
| HF | 0.10 | -0.41 | 0.39 | 0.30 | -0.02 | -0.05 | 0.03 | 0.03 | 0.20 | 0.11 | -0.03 | -0.20 | 4.2 | 0.37 |
| VHF | -0.10 | -0.45 | 0.41 | 0.28 | 0.14 | -0.06 | 0.08 | -0.01 | 0.18 | -0.09 | -0.09 | 0.01 | 3.8 | 0.49 |
| LFHF | 0.13 | 0.58 | 0.12 | 0.46 | 0.05 | -0.06 | -0.01 | 0.04 | -0.18 | -0.14 | 0.02 | 0.06 | 2.6 | 0.28 |
| LFn | 0.08 | 0.51 | 0.31 | 0.71 | 0.00 | -0.02 | -0.03 | 0.11 | -0.13 | -0.05 | 0.01 | -0.02 | 2.4 | 0.12 |
| HFn | -0.15 | -0.63 | 0.06 | -0.24 | -0.10 | 0.00 | 0.03 | 0.07 | 0.15 | 0.22 | -0.11 | -0.13 | 2.2 | 0.28 |
| LnHF | 0.00 | -0.60 | 0.38 | 0.33 | -0.12 | 0.01 | 0.00 | 0.12 | 0.18 | -0.01 | 0.06 | -0.22 | 3.1 | 0.13 |
| SD2 | 0.85 | 0.12 | -0.06 | 0.02 | 0.02 | -0.10 | -0.05 | 0.00 | 0.03 | 0.09 | 0.30 | 0.03 | 1.4 | 0.10 |
| SD1SD2 | -0.02 | -1.16 | -0.11 | 0.03 | 0.02 | -0.19 | -0.02 | 0.05 | -0.13 | -0.52 | 0.03 | 0.15 | 1.6 | 0.02 |
| S | 0.54 | -0.05 | -0.02 | -0.11 | -0.07 | 0.02 | 0.06 | 0.00 | 0.18 | 0.03 | 0.51 | -0.07 | 2.4 | 0.14 |
| CSI | 0.01 | 0.88 | 0.03 | -0.18 | -0.12 | -0.11 | 0.00 | 0.04 | 0.28 | 0.10 | -0.04 | 0.13 | 1.4 | 0.12 |
| CVI | 0.45 | -0.35 | -0.12 | 0.00 | 0.02 | 0.01 | -0.03 | 0.00 | 0.08 | 0.09 | 0.41 | 0.27 | 4.0 | 0.01 |
| CSI_Modified | 0.23 | 0.34 | -0.16 | 0.01 | -0.11 | 0.02 | 0.11 | -0.01 | 0.07 | -0.01 | -0.08 | 0.68 | 2.1 | 0.17 |
| PIP | -0.01 | 0.01 | 0.00 | -0.06 | 1.04 | 0.05 | 0.02 | 0.03 | 0.01 | -0.08 | -0.02 | -0.02 | 1.0 | 0.00 |
| IALS | 0.01 | 0.02 | 0.01 | -0.01 | 1.02 | 0.07 | 0.00 | 0.04 | -0.04 | 0.00 | -0.03 | -0.06 | 1.0 | 0.02 |
| PSS | 0.01 | 0.11 | 0.12 | -0.03 | 1.07 | 0.22 | 0.03 | 0.03 | 0.02 | -0.07 | 0.02 | -0.04 | 1.1 | 0.13 |
| PAS | 0.03 | 0.05 | -0.17 | 0.05 | 0.70 | -0.17 | 0.05 | 0.06 | -0.02 | 0.01 | 0.03 | -0.03 | 1.3 | 0.19 |
| GI | -0.05 | 0.03 | 0.00 | -0.18 | 0.01 | -0.14 | 0.08 | 0.92 | -0.02 | -0.05 | 0.02 | 0.04 | 1.2 | 0.08 |
| SI | 0.03 | 0.10 | -0.04 | -0.11 | -0.03 | 0.03 | -0.22 | 0.69 | -0.02 | -0.10 | -0.04 | 0.01 | 1.4 | 0.31 |
| AI | 0.00 | -0.12 | -0.04 | -0.15 | 0.17 | 0.04 | 0.01 | 0.72 | -0.07 | 0.21 | -0.02 | -0.07 | 1.5 | 0.31 |
| PI | 0.16 | 0.07 | -0.12 | 0.15 | 0.02 | 0.01 | 0.62 | -0.04 | -0.14 | 0.18 | -0.06 | -0.10 | 1.8 | 0.35 |
| C1a | 0.01 | 0.21 | 0.03 | 0.14 | 0.10 | -0.02 | -0.58 | -0.06 | -0.06 | 0.00 | -0.06 | 0.05 | 1.6 | 0.55 |
| C2a | -0.05 | 0.00 | 0.10 | -0.08 | 0.08 | -0.09 | 0.99 | -0.01 | -0.04 | -0.07 | 0.02 | 0.08 | 1.1 | 0.05 |
| Ca | -0.10 | 0.17 | 0.08 | -0.02 | 0.06 | 0.00 | 0.88 | -0.06 | 0.00 | -0.07 | 0.03 | 0.18 | 1.2 | 0.27 |
| DFA_alpha1 | -0.04 | 0.79 | 0.26 | 0.32 | -0.17 | 0.02 | 0.06 | 0.03 | -0.08 | -0.02 | -0.03 | 0.08 | 1.7 | 0.07 |
| DFA_alpha1_ExpRange | 0.09 | -0.09 | 0.46 | -0.16 | -0.07 | 0.09 | 0.12 | 0.04 | -0.29 | 0.07 | -0.03 | -0.09 | 2.8 | 0.51 |
| DFA_alpha1_ExpMean | -0.14 | 0.37 | 0.77 | 0.09 | -0.11 | 0.08 | 0.08 | 0.07 | -0.26 | 0.15 | -0.06 | 0.03 | 2.1 | 0.31 |
| DFA_alpha1_DimRange | -0.18 | 0.06 | 0.01 | -0.11 | -0.01 | 0.01 | 0.04 | 0.02 | -0.41 | -0.07 | -0.03 | -0.03 | 1.7 | 0.79 |
| DFA_alpha1_DimMean | -0.10 | 0.28 | 0.17 | 0.38 | 0.13 | -0.19 | -0.06 | 0.06 | 0.12 | 0.27 | -0.02 | 0.08 | 5.0 | 0.43 |
| DFA_alpha2 | -0.08 | 0.64 | 0.01 | -0.06 | 0.19 | -0.01 | -0.04 | -0.05 | 0.08 | 0.19 | -0.05 | 0.05 | 1.5 | 0.53 |
| DFA_alpha2_ExpRange | -0.11 | 0.00 | -0.10 | 0.65 | -0.06 | 0.10 | -0.03 | -0.22 | 0.03 | 0.01 | -0.04 | -0.02 | 1.4 | 0.46 |
| DFA_alpha2_ExpMean | -0.01 | 0.47 | -0.07 | 0.08 | 0.13 | 0.00 | 0.07 | 0.05 | 0.30 | 0.04 | -0.01 | 0.04 | 2.1 | 0.64 |
| DFA_alpha2_DimRange | -0.01 | 0.10 | -0.01 | 0.59 | -0.01 | 0.01 | -0.07 | -0.13 | 0.08 | 0.06 | -0.08 | -0.06 | 1.3 | 0.52 |
| DFA_alpha2_DimMean | -0.03 | 0.01 | 0.21 | -0.29 | 0.03 | -0.08 | -0.04 | -0.08 | -0.10 | 0.10 | -0.03 | -0.03 | 3.0 | 0.85 |
| ApEn | 0.09 | -0.03 | 0.55 | -0.72 | 0.02 | -0.13 | 0.02 | 0.01 | -0.05 | -0.35 | 0.08 | -0.07 | 2.6 | 0.13 |
| SampEn | 0.06 | -0.01 | 0.89 | -0.24 | 0.13 | -0.05 | -0.04 | -0.04 | 0.09 | -0.40 | -0.01 | -0.19 | 1.8 | 0.05 |
| ShanEn | 0.20 | -0.08 | 0.17 | -0.25 | -0.28 | 0.17 | -0.05 | 0.07 | -0.01 | 0.29 | -0.03 | 0.12 | 6.0 | 0.37 |
| FuzzyEn | 0.06 | -0.17 | 0.90 | -0.20 | 0.04 | -0.10 | -0.06 | -0.08 | 0.01 | -0.24 | -0.03 | -0.08 | 1.4 | 0.04 |
| MSE | 0.06 | 0.08 | -0.05 | -0.63 | -0.06 | 0.19 | -0.07 | 0.10 | 0.06 | -0.10 | 0.04 | -0.08 | 1.5 | 0.39 |
| CMSE | -0.10 | 0.38 | 0.35 | -0.23 | -0.14 | 0.11 | 0.02 | -0.06 | 0.59 | -0.14 | 0.08 | -0.04 | 3.4 | 0.16 |
| RCMSE | -0.08 | 0.43 | 0.38 | -0.06 | -0.12 | 0.11 | 0.03 | -0.04 | 0.59 | -0.10 | 0.05 | -0.02 | 3.0 | 0.13 |
| CD | 0.03 | 0.26 | 0.87 | -0.05 | 0.04 | -0.12 | 0.02 | -0.02 | 0.19 | 0.06 | -0.14 | 0.00 | 1.4 | 0.17 |
| HFD | -0.16 | -0.73 | 0.18 | -0.19 | 0.04 | 0.05 | -0.07 | -0.05 | 0.04 | 0.00 | 0.00 | 0.07 | 1.4 | 0.23 |
| KFD | 0.01 | -0.29 | 0.50 | -0.43 | 0.06 | -0.13 | -0.07 | -0.01 | 0.12 | 0.09 | 0.07 | 0.11 | 3.3 | 0.32 |
| LZC | -0.06 | -0.30 | 0.81 | 0.11 | 0.03 | 0.00 | 0.00 | -0.05 | -0.06 | -0.15 | 0.17 | 0.04 | 1.5 | 0.18 |
# insight::display(efa, threshold="max", sort=TRUE)
plot(efa, size_text = 3) +
theme(axis.text.y=element_text(size=5))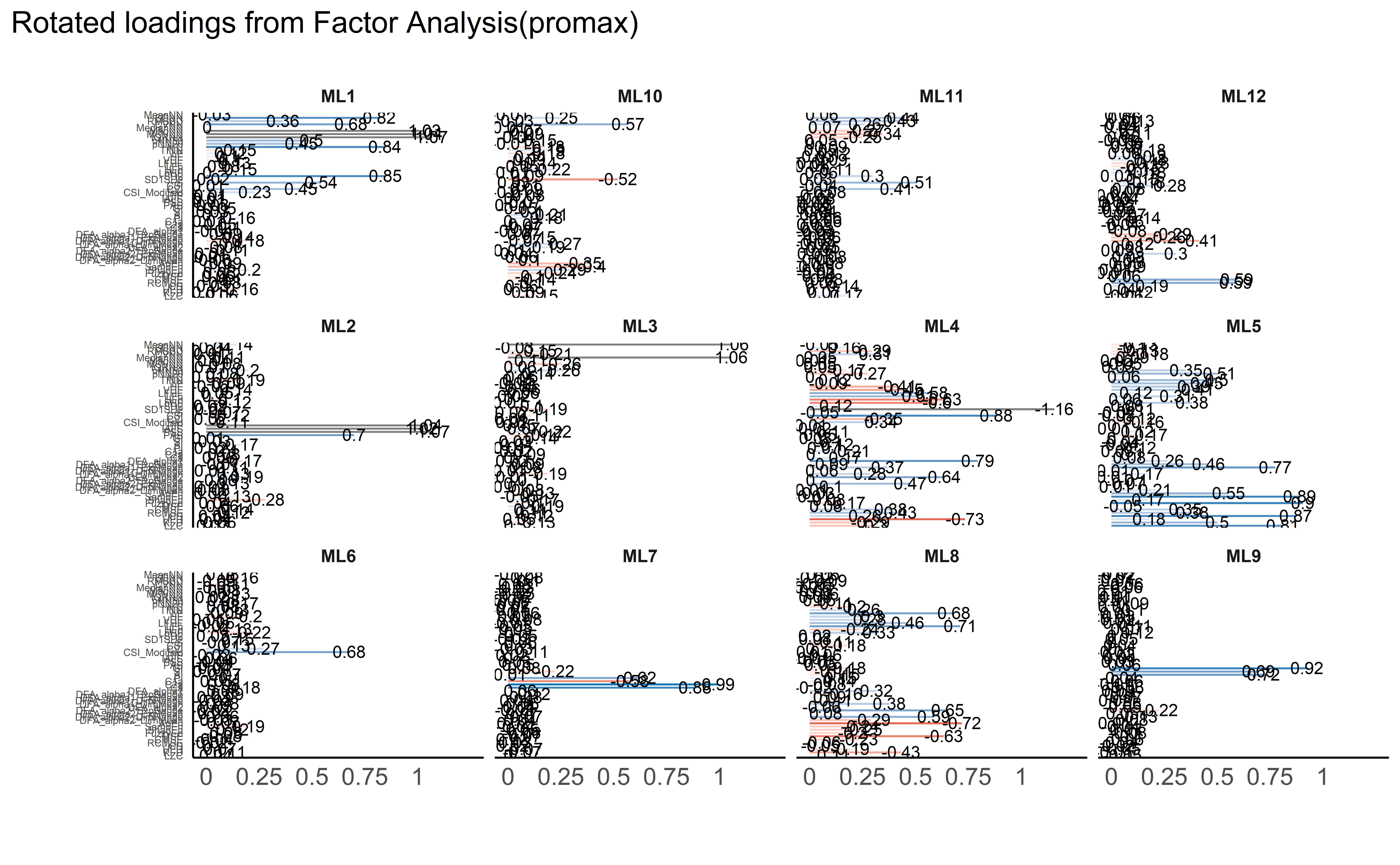
Figure 4: EFA 12-factor Rotated Loadings
table_clusters$EFA_12 <- as.character(parameters::closest_component(efa))Cluster Structure
Clustering
Clustering is the process of assigning data to different groupings according to their similarity. Each approach under the clustering group will have different set of criteria to determine the association between data points. However, they rely on the same set of distance and linkage methods where the formal determines how the distance (similarity) between two points are calculated and the later determines how inter-cluster distance can be derived.
In this paper, two non-hierarchical clustering methods employed are k-means and k-medoids in which data points are partitioned into k sets (clusters). The optimal k value is determined by the n_factor function from the parameters package where the solution with the maximum consensus of a large number of methods is chosen as the final outcome. The hierarchical clustering relied on euclidean distance method (shortest distance between two data points) and average linkage method (average of all inter-cluster’s distances while compensating for the number of points in each cluster) to partition the data into clusters.
For the density-based clustering, the epsilon value (maximum distance between two points to be in assigned to the same cluster) in DBSCAN clustering is determined by the n_clusters_dbscan function in parameters package. The minimum number of points of a cluster is set to 2 for both DBSCAN and HDBSCAN.
Last but not least, the model-based clustering, based on finite Gaussian mixture modelling, was applied to the dataset to estimate the probability of each data being assigned to different cluster solutions. An advantage of this method is that it automatically identifies the optimal number of clusters and thus no parameters were required as pre-requisites.
# Preprocessing
z <- effectsize::standardize(data[hrv_cols])
z <- datawizard::data_transpose(z)
z <- z[ , colSums(is.na(z)) < 0.2 * nrow(z)] # keep only participants with few nans
z[is.na(z)] <- 0 # mean imputationK-Means
How Many Clusters
set.seed(3)
n <- parameters::n_clusters(z,
standardize = FALSE,
package = "all",
nbclust_method = "average",
fast = FALSE)
n> # Method Agreement Procedure:
>
> The choice of 10 clusters is supported by 6 (24.00%) methods out of 25 (Gap_Dudoit2002, DB, SDindex, SDbw, gamma, gplus).plot(n)
2-K Solution
set.seed(3)
rez <- parameters::cluster_analysis(z, standardize = FALSE, n = 2, method = "hkmeans")
as.data.frame(rez)[1:3]> Cluster n_Obs Sum_Squares
> 1 1 27 5053
> 2 2 30 7649insight::display(parameters::cluster_performance(rez))| Sum_Squares_Total | Sum_Squares_Between | Sum_Squares_Within | R2 |
|---|---|---|---|
| 15329.70 | 2628.11 | 12701.60 | 0.17 |
plot(rez)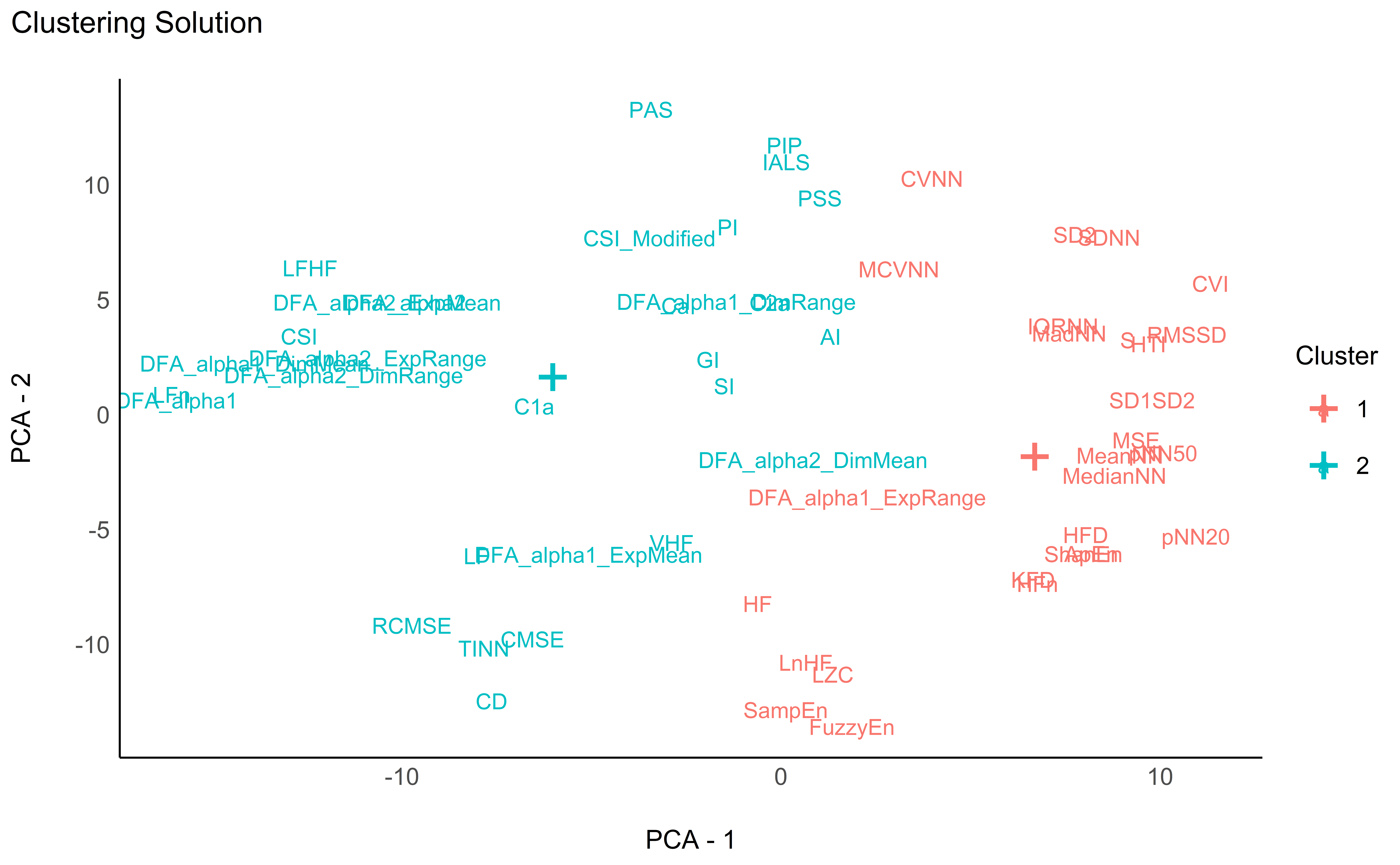
Figure 5: K-Means 2-cluster Clustegram solution
table_clusters <- assign_clusters(table_clusters, z, clusters = as.character(predict(rez)), col = "Kmeans_2")7-K Solution
set.seed(3)
rez <- parameters::cluster_analysis(z, standardize = FALSE, n = 7, method = "hkmeans")
as.data.frame(rez)[1:3]> Cluster n_Obs Sum_Squares
> 1 1 12 1790
> 2 2 10 1017
> 3 3 12 1732
> 4 4 11 1875
> 5 5 4 164
> 6 6 5 852
> 7 7 3 203insight::display(parameters::cluster_performance(rez))| Sum_Squares_Total | Sum_Squares_Between | Sum_Squares_Within | R2 |
|---|---|---|---|
| 15329.70 | 7696.84 | 7632.86 | 0.50 |
plot(rez)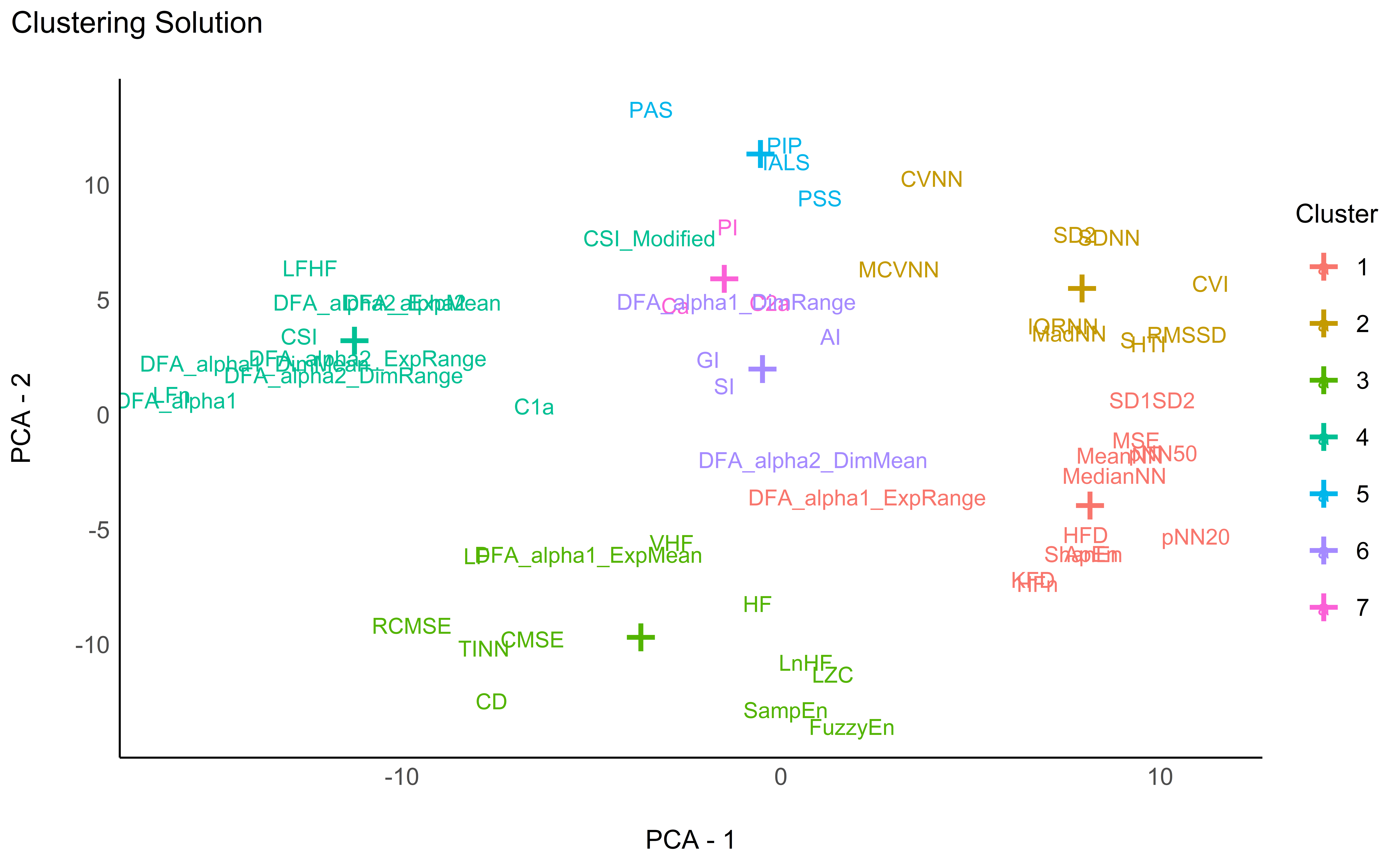
Figure 6: K-Means 7-cluster Clustegram solution
table_clusters <- assign_clusters(table_clusters, z, clusters = as.character(predict(rez)), col = "Kmeans_7")10-K Solution
set.seed(3)
rez <- parameters::cluster_analysis(z, n = 10, method = "hkmeans")
as.data.frame(rez)[1:3]> Cluster n_Obs Sum_Squares
> 1 1 5 660
> 2 2 12 1327
> 3 3 12 1975
> 4 4 6 850
> 5 5 6 968
> 6 6 5 877
> 7 7 4 188
> 8 8 3 264
> 9 9 3 217
> 10 10 1 0insight::display(parameters::cluster_performance(rez))| Sum_Squares_Total | Sum_Squares_Between | Sum_Squares_Within | R2 |
|---|---|---|---|
| 16856.00 | 9529.66 | 7326.34 | 0.57 |
plot(rez)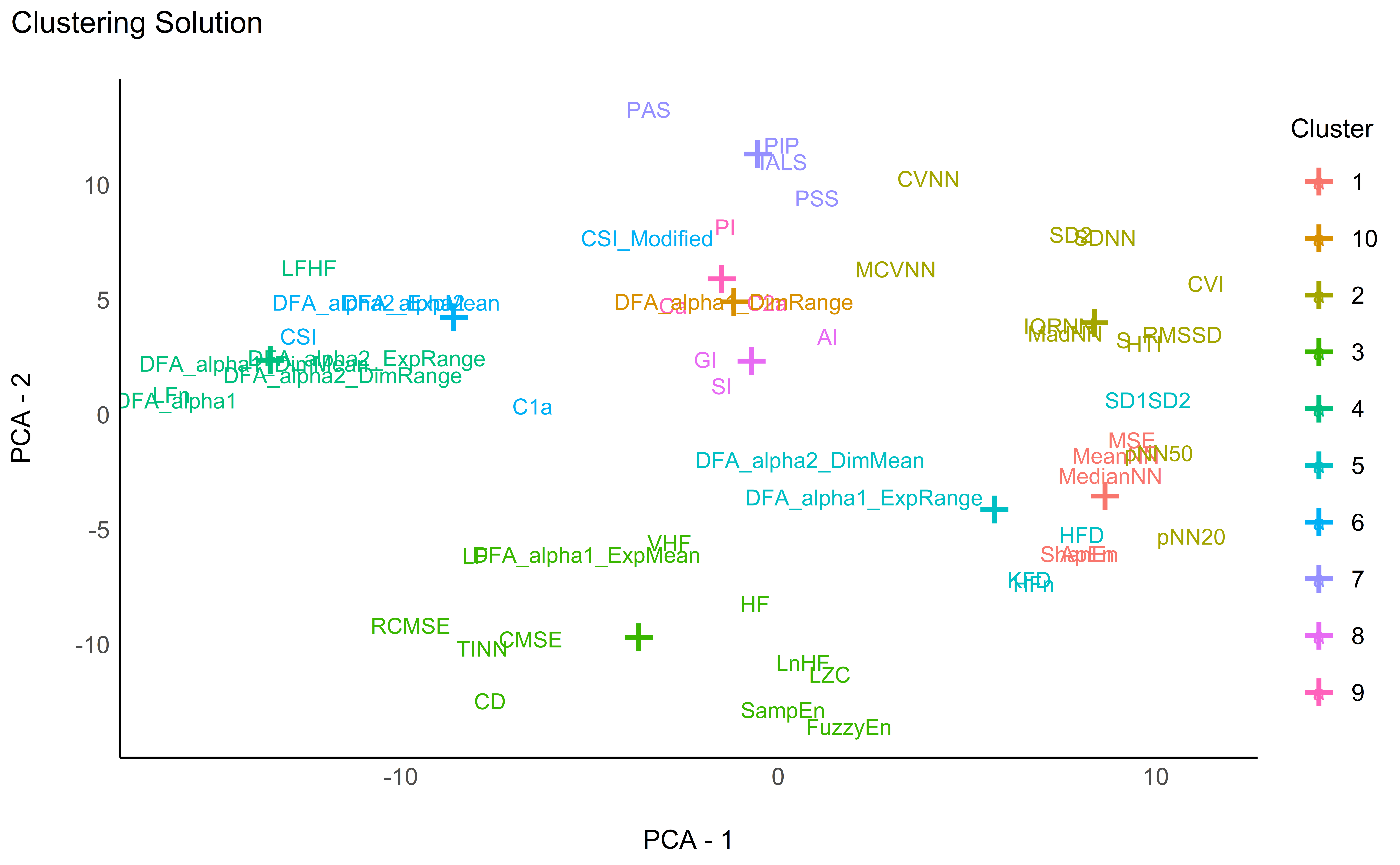
Figure 7: K-Means 10-cluster Clustegram solution
table_clusters <- assign_clusters(table_clusters, z, clusters = as.character(predict(rez)), col = "Kmeans_10")K-meloid
set.seed(3)
rez <- parameters::cluster_analysis(z, method = "pamk", distance_method = "euclidean")
as.data.frame(rez)[1:3]> Cluster n_Obs Sum_Squares
> 1 1 24 5745
> 2 2 18 3536
> 3 3 15 3406insight::display(parameters::cluster_performance(rez))| Sum_Squares_Total | Sum_Squares_Between | Sum_Squares_Within | R2 |
|---|---|---|---|
| 16856.00 | 4169.19 | 12686.81 | 0.25 |
plot(rez)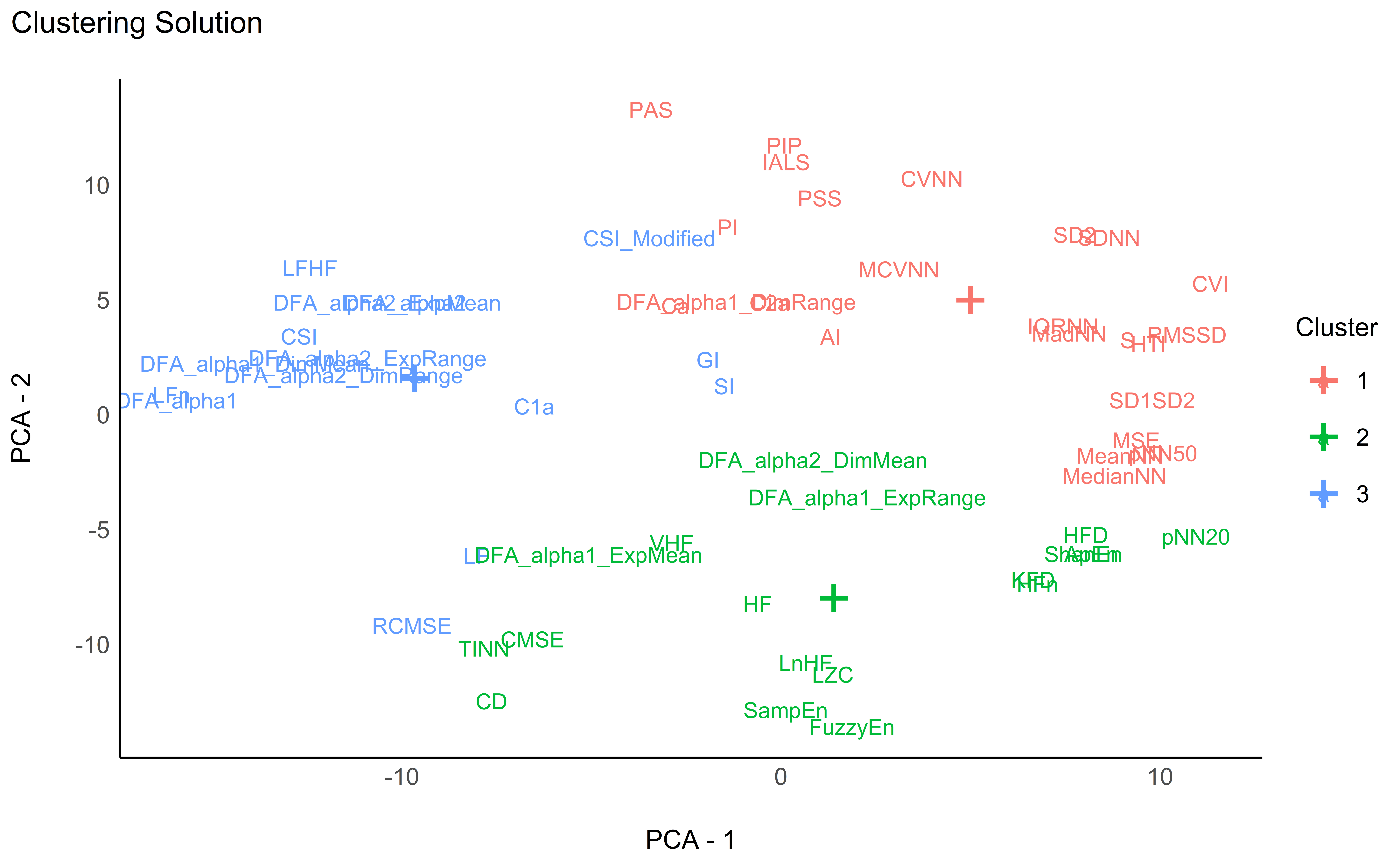
Figure 8: K-Meloid 3-cluster Clustegram solution
table_clusters <- assign_clusters(table_clusters, z, clusters = as.character(predict(rez)), col = "Kmeloid")Hierarchical Clustering
Clustering (95%)
set.seed(3)
rez <- parameters::cluster_analysis(z, n = NULL, standardize = FALSE, method = "hclust", distance_method = "euclidean", hclust_method = "average", ci = 0.95, iterations = 2000)
as.data.frame(rez)[1:3]> Cluster n_Obs Sum_Squares
> 1 0 32 8658.3
> 2 1 2 6.1
> 3 10 2 109.7
> 4 11 3 244.5
> 5 2 2 11.8
> 6 3 2 21.8
> 7 4 2 34.7
> 8 5 2 53.7
> 9 6 2 70.9
> 10 7 4 164.0
> 11 8 2 78.1
> 12 9 2 108.6insight::display(parameters::cluster_performance(rez))| Sum_Squares_Total | Sum_Squares_Between | Sum_Squares_Within | R2 |
|---|---|---|---|
| 15329.70 | 5767.45 | 903.92 | 0.38 |
plot(rez)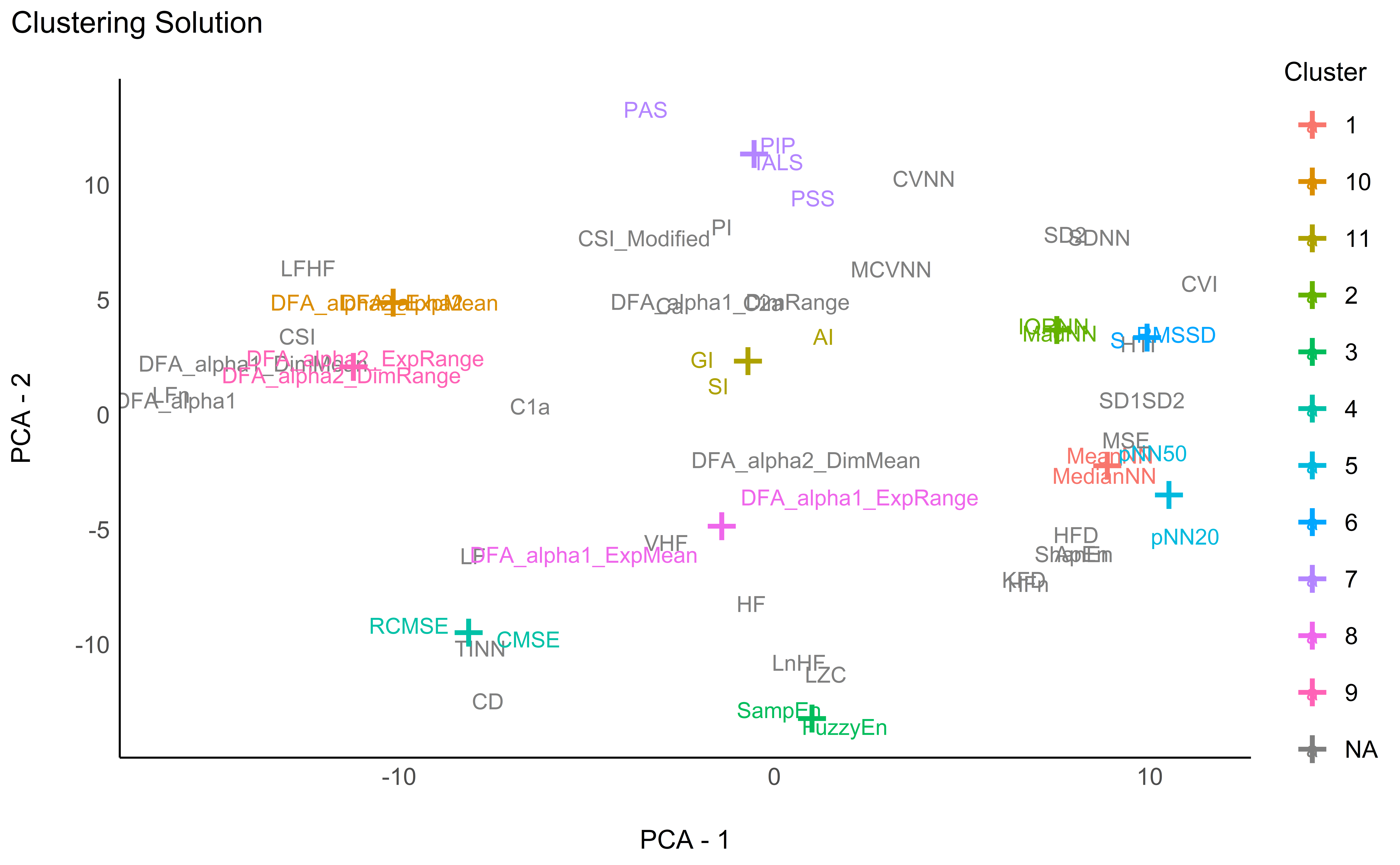
Figure 9: Hierarchical Clustering 11-cluster dendogram solution
table_clusters <- assign_clusters(table_clusters, z, clusters = as.character(predict(rez)), col = "Hclust_95")clusters <- table_clusters$Hclust_95
names(clusters) <- table_clusters$Index
library(ggraph)
plot_dendrogram(model = attributes(rez)$model$hclust, clusters = clusters, ylim = -10, nudge_y = -0.05, angle = 90)
Clustering (90%)
set.seed(3)
rez <- parameters::cluster_analysis(z, n = NULL, standardize = FALSE, method = "hclust", distance_method = "euclidean", hclust_method = "average", ci = 0.90, iterations = 2000)
as.data.frame(rez)[1:3]> Cluster n_Obs Sum_Squares
> 1 0 24 6177.4
> 2 1 2 6.1
> 3 10 2 108.6
> 4 11 2 109.7
> 5 12 3 202.6
> 6 13 3 244.5
> 7 2 2 21.8
> 8 3 2 34.7
> 9 4 2 53.7
> 10 5 2 70.9
> 11 6 4 173.0
> 12 7 4 164.0
> 13 8 2 78.1
> 14 9 3 157.6insight::display(parameters::cluster_performance(rez))| Sum_Squares_Total | Sum_Squares_Between | Sum_Squares_Within | R2 |
|---|---|---|---|
| 15329.70 | 7727.04 | 1425.29 | 0.50 |
plot(rez)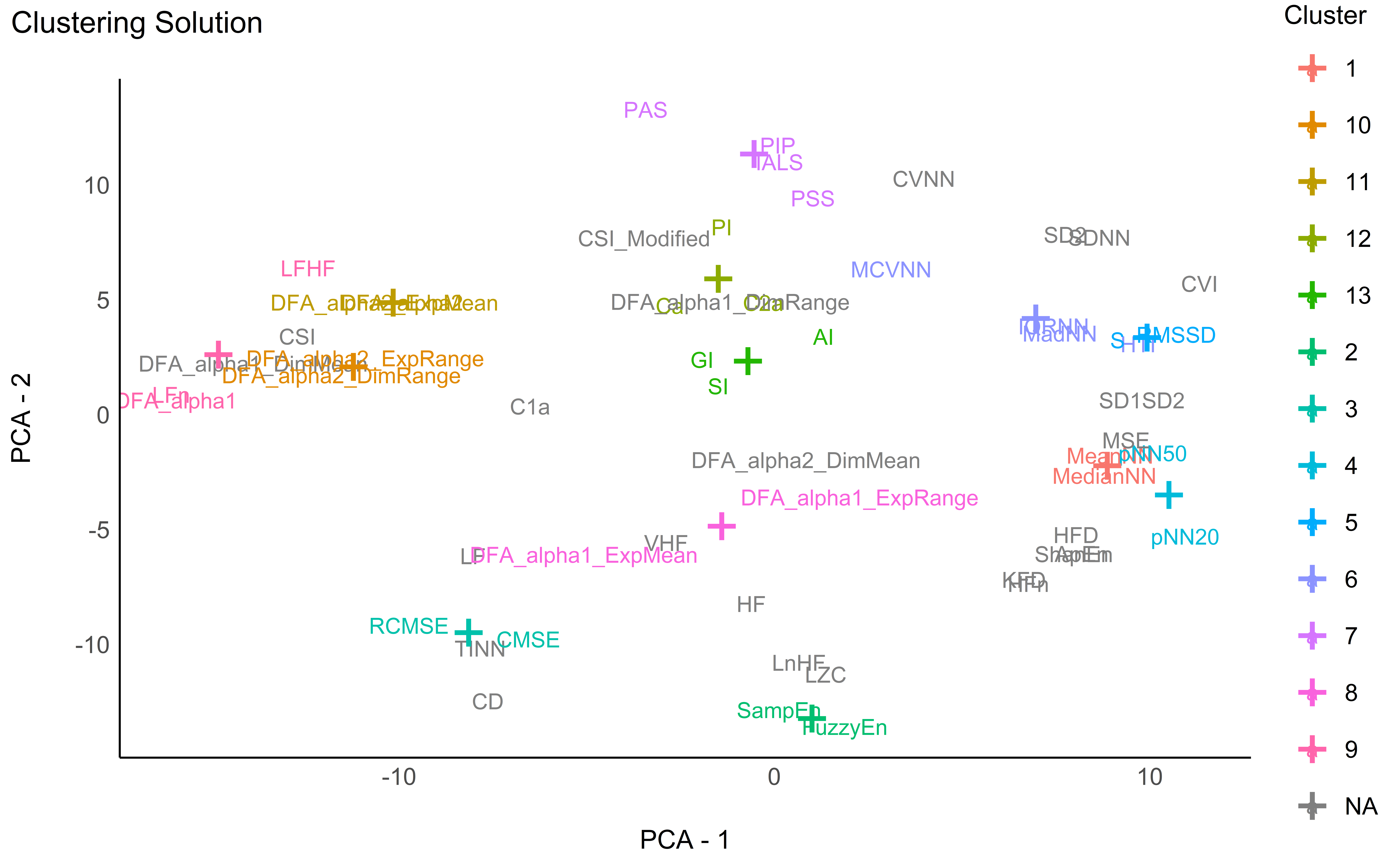
Figure 10: Hierarchical Clustering 13-cluster dendogram solution
table_clusters <- assign_clusters(table_clusters, z, clusters = as.character(predict(rez)), col = "Hclust_90")clusters <- table_clusters$Hclust_90
names(clusters) <- table_clusters$Index
plot_dendrogram(model = attributes(rez)$model$hclust, clusters = clusters, ylim = -10, nudge_y = -0.05, angle = 90,)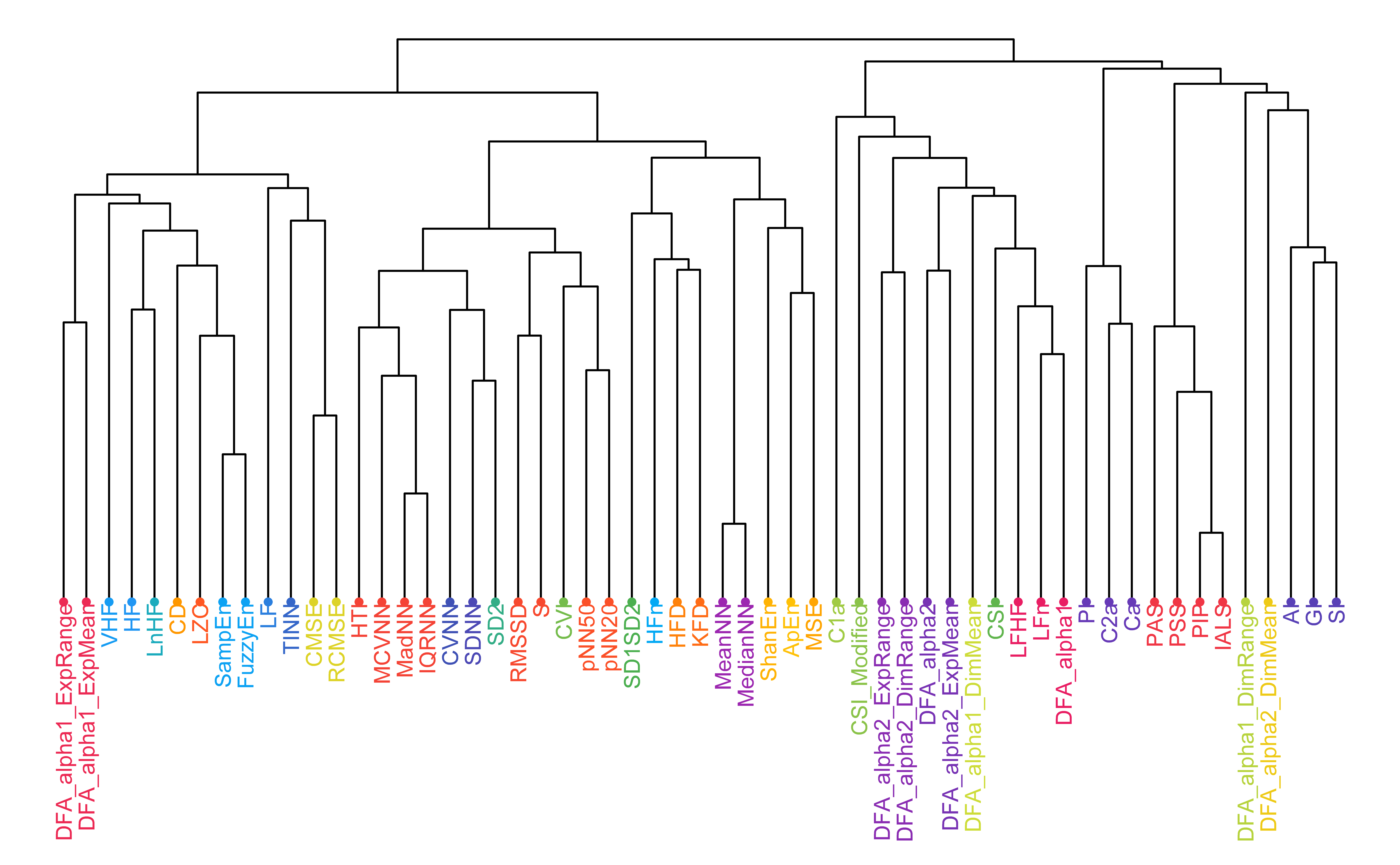
DBSCAN
Parameters Selection
n <- n_clusters_dbscan(z, standardize = FALSE, eps_range = c(0.1, 30), min_size = 2, method = "kNN")
plot(n)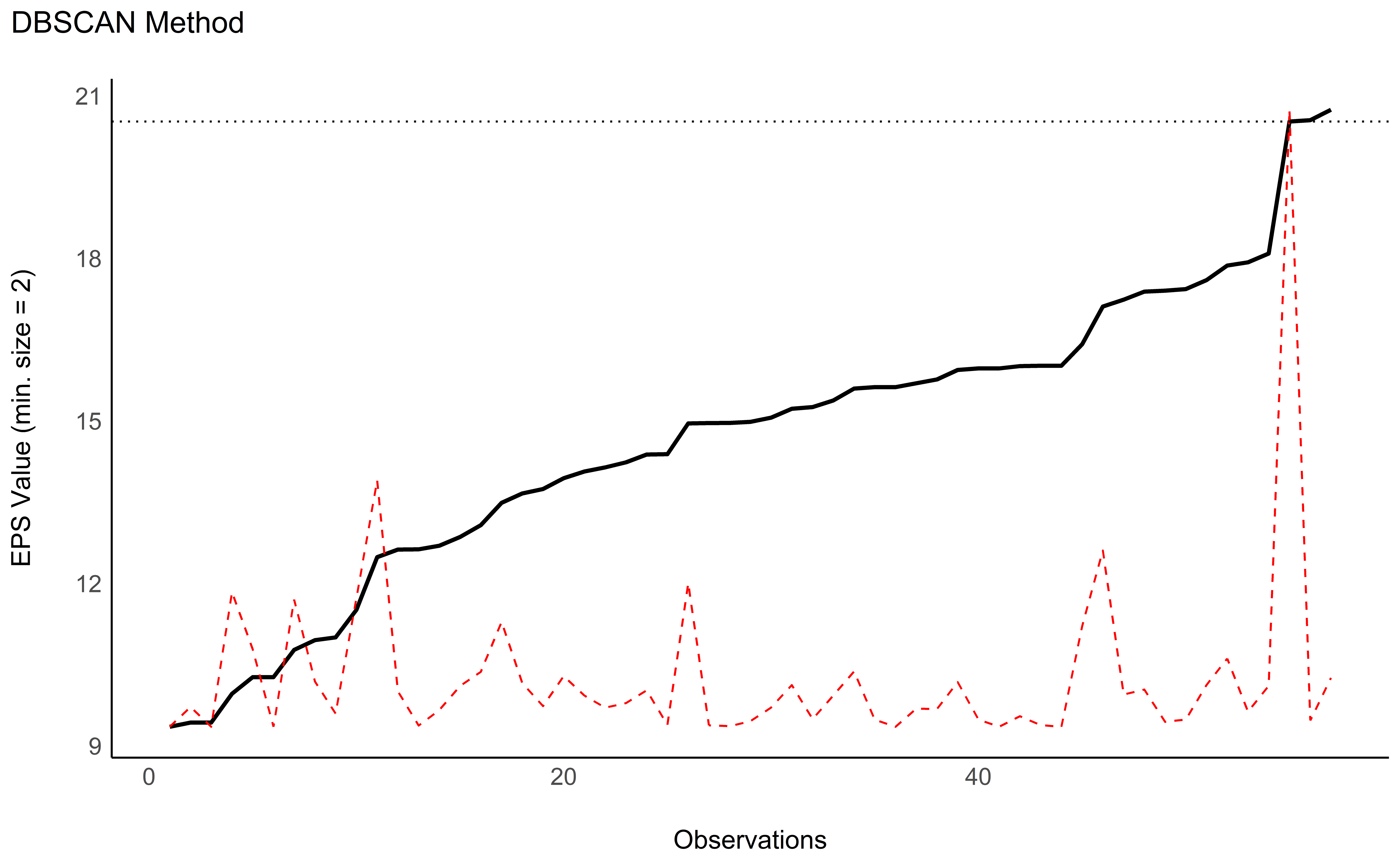
DBSCAN
rez <- parameters::cluster_analysis(z, n = NULL, standardize = FALSE, method = "dbscan", dbscan_eps = 10, min_size = 2, borderPoints = TRUE)
as.data.frame(rez)[1:3]> Cluster n_Obs Sum_Squares
> 1 0 43 11500.3
> 2 1 2 6.1
> 3 2 2 48.9
> 4 3 3 76.1
> 5 4 3 62.0
> 6 5 2 21.8
> 7 6 2 34.7insight::display(parameters::cluster_performance(rez))| Sum_Squares_Total | Sum_Squares_Between | Sum_Squares_Within | R2 |
|---|---|---|---|
| 15329.70 | 3579.75 | 249.67 | 0.23 |
plot(rez)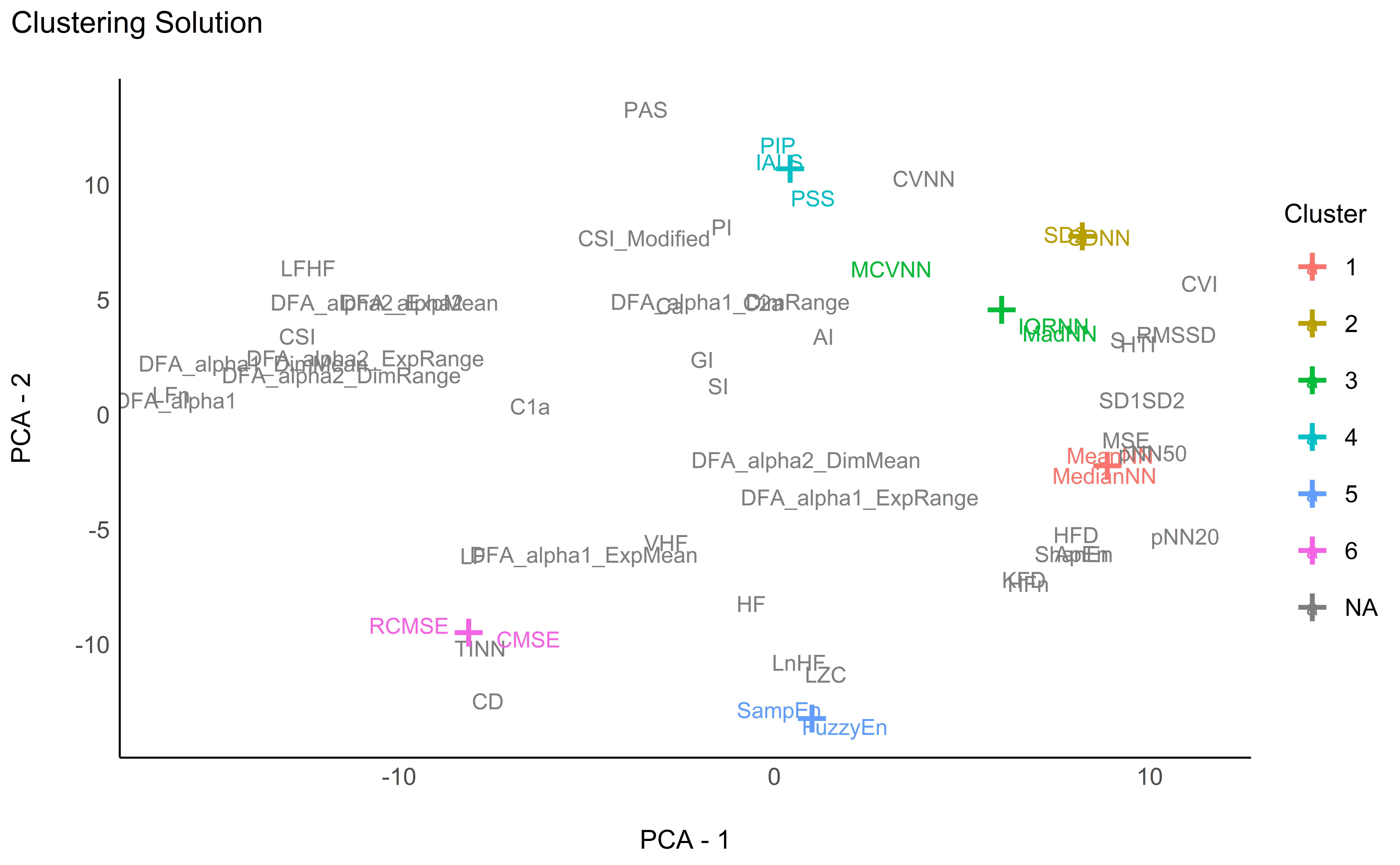
Figure 11: DBSCAN 6-cluster clustergram solution
table_clusters <- assign_clusters(table_clusters, z, clusters = as.character(predict(rez)), col = "DBSCAN")HDBSCAN
rez <- parameters::cluster_analysis(z, n = NULL, standardize = FALSE, method = "hdbscan", min_size = 2)
as.data.frame(rez)[1:3]> Cluster n_Obs Sum_Squares
> 1 0 8 1983.8
> 2 1 4 164.0
> 3 10 8 1127.3
> 4 11 2 70.9
> 5 12 3 147.9
> 6 13 4 173.0
> 7 14 2 53.7
> 8 15 2 6.1
> 9 2 3 244.5
> 10 3 3 202.6
> 11 4 2 109.7
> 12 5 2 108.6
> 13 6 6 758.2
> 14 7 4 401.5
> 15 8 2 78.1
> 16 9 2 34.7insight::display(parameters::cluster_performance(rez))| Sum_Squares_Total | Sum_Squares_Between | Sum_Squares_Within | R2 |
|---|---|---|---|
| 15329.70 | 9665.19 | 3680.69 | 0.63 |
plot(rez)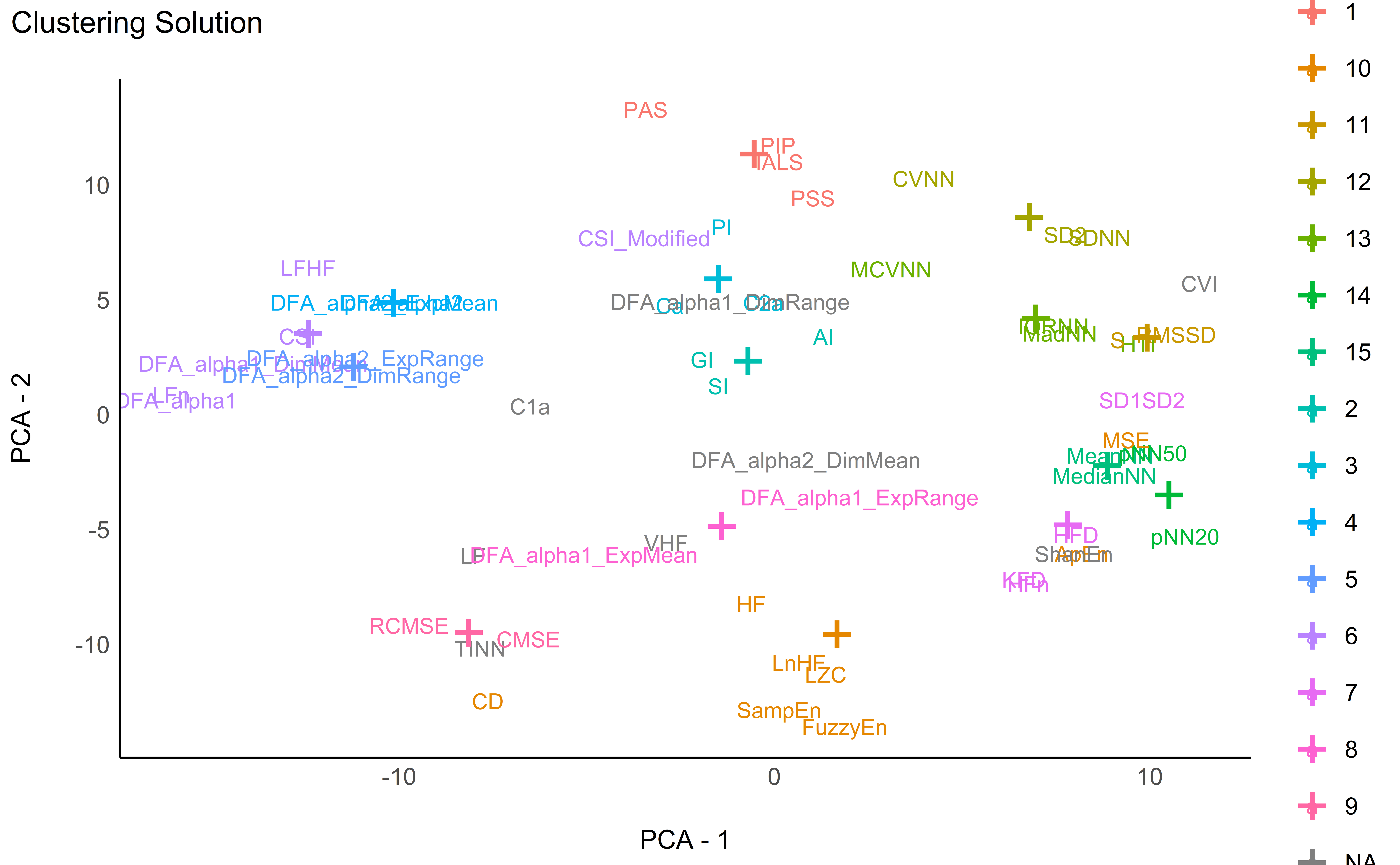
Figure 12: HDBSCAN 13-cluster clustergram solution
table_clusters <- assign_clusters(table_clusters, z, clusters = as.character(predict(rez)), col = "HDBSCAN")Mixture Modelling
suppressMessages(library(mclust))
library(mclust)
rez <- parameters::cluster_analysis(z, method = "mixture")
as.data.frame(rez)[1:3]> Cluster n_Obs Sum_Squares
> 1 1 10 2260
> 2 2 12 1327
> 3 3 17 3341
> 4 4 11 2146
> 5 5 4 188
> 6 6 3 217insight::display(parameters::cluster_performance(rez))| Sum_Squares_Total | Sum_Squares_Between | Sum_Squares_Within | R2 |
|---|---|---|---|
| 16856.00 | 7376.66 | 9479.34 | 0.44 |
plot(rez)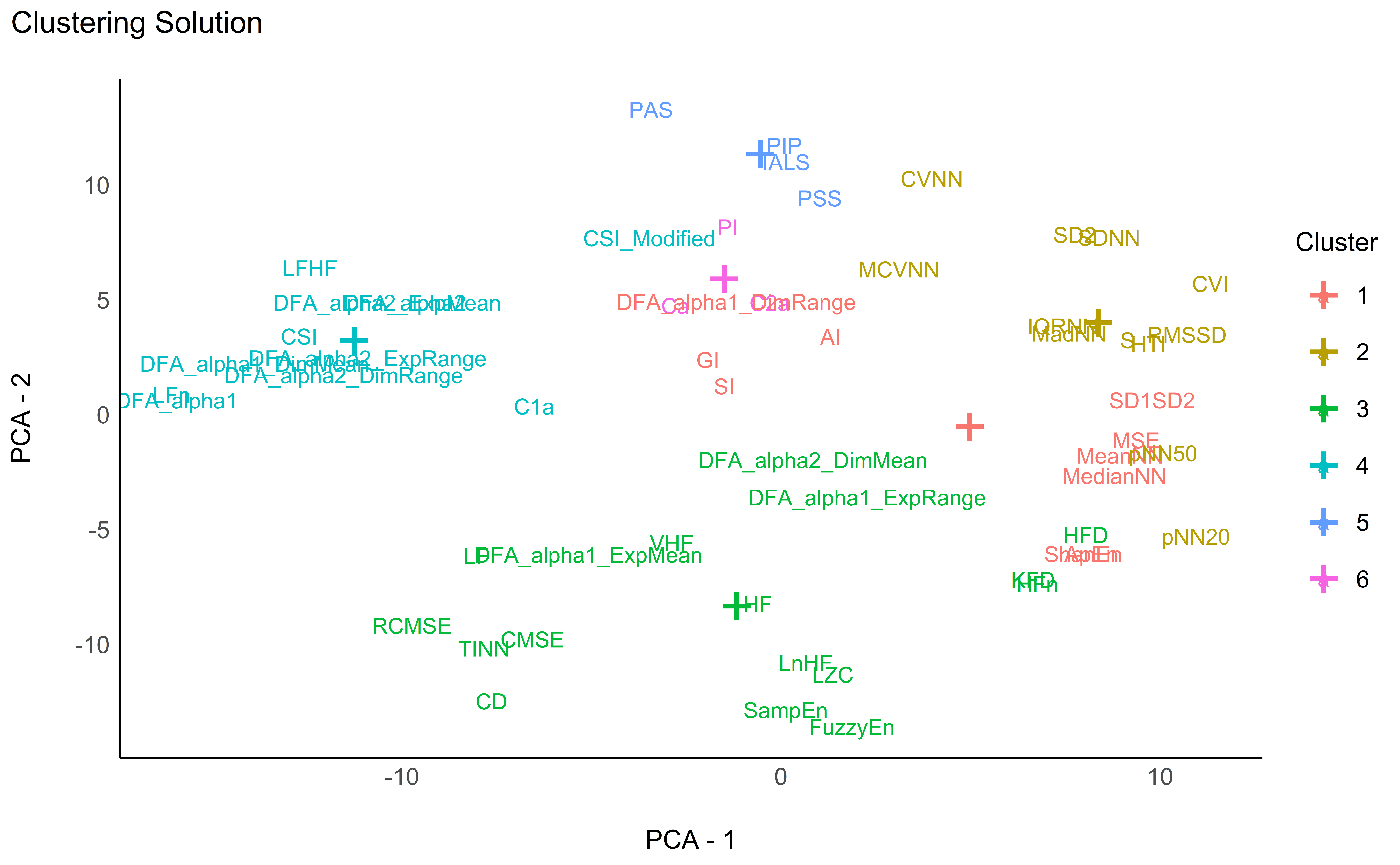
Figure 13: Mixture Modelling 6-cluster clustergram solution
table_clusters <- assign_clusters(table_clusters, z, clusters = as.character(predict(rez)), col = "Mixture")Network Structure
Network
EGA
Traditionally, PCA and FA are the two most common factor analytic methods that were used to estimate the number of underlying factors in the observed data. EGA is a newly introduced method which applies the network modeling framework to estimate a network based on the relationships between the variables and subsequently identify the clusters of the variables in the network (community detection algorithms: louvain or walktrap). Simulation data has not only shown that EGA has higher accuracy than most PCA and FA methods.
In this paper, GLASSO and TMFG were used to estimate two separate networks or two partial correlation matrices. The louvian algorithm is then used to find the number of clusters based on the correlation matrices.
# Preprocessing
z <- effectsize::standardize(data[hrv_cols])
z <- z[rowSums(is.na(z)) < 0.2 * ncol(z), ] # keep only participants with few nans
z[is.na(z)] <- 0 EGA glasso
suppressMessages(library(EGAnet))
# Run EGA
ega_glasso <- EGAnet::EGA(z, model = "glasso", algorithm = "louvain", plot.EGA = TRUE)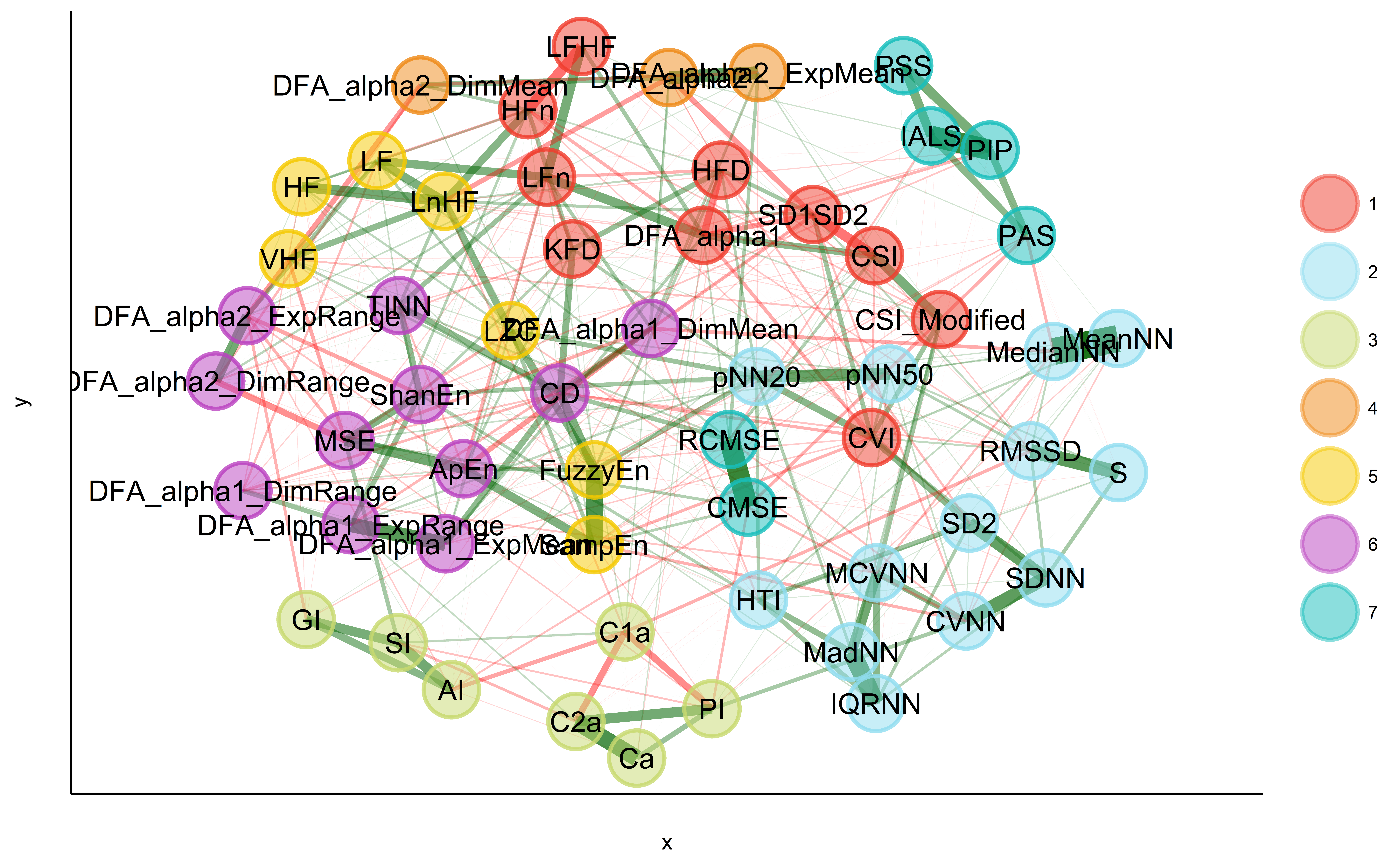
Figure 14: EGA with GLASSO 7-cluster network solution
EGA TMFG
# Run EGA
ega_TMFG <- EGAnet::EGA(z, model = "TMFG", algorithm = "louvain", plot.EGA = TRUE)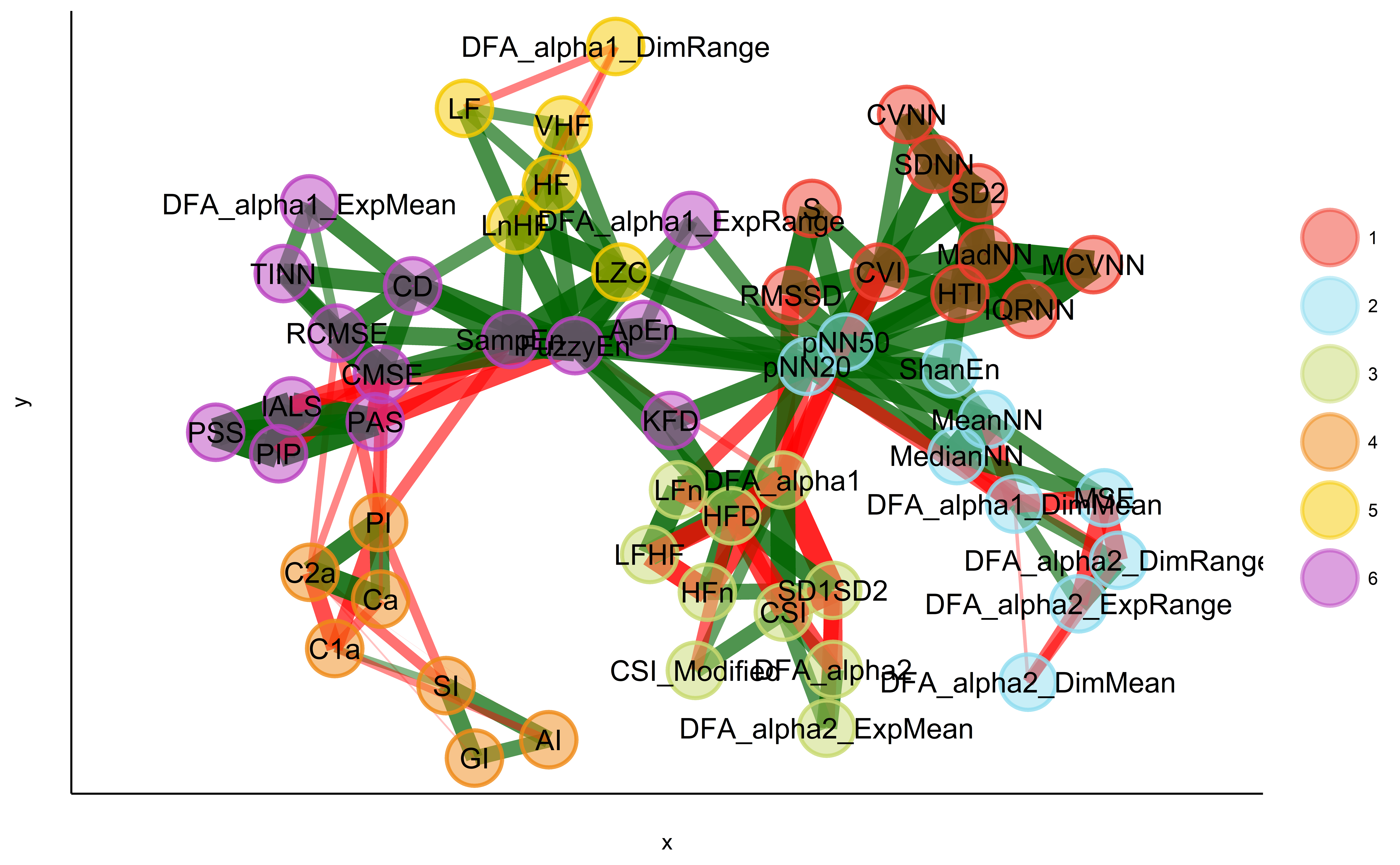
Figure 15: EGA with TMFG 6-cluster network solution
table_clusters$EGA_glasso <- ega_glasso$dim.variables[table_clusters$Index, "dimension"]
table_clusters$EGA_TMFG <- ega_TMFG$dim.variables[table_clusters$Index, "dimension"]Centrality
graph_glasso <- EGAnet::ega_to_tidygraph(ega_glasso) #from Dom's PR
graph_glasso <- tidygraph::tbl_graph(nodes = graph_glasso$nodes, edges = graph_glasso$edges, directed = FALSE) %>%
tidygraph::activate(nodes) %>%
mutate(centrality = tidygraph::centrality_degree())
as.list(graph_glasso)$nodes %>%
group_by(dimension) %>%
arrange(desc(centrality)) %>%
slice_head(n=1) %>%
knitr::kable()| name | dimension | centrality |
|---|---|---|
| LFn | 1 | 24 |
| pNN20 | 2 | 28 |
| SI | 3 | 12 |
| DFA_alpha2 | 4 | 12 |
| LnHF | 5 | 22 |
| CD | 6 | 28 |
| RCMSE | 7 | 20 |
graph_TMFG <- EGAnet::ega_to_tidygraph(ega_TMFG) #from Dom's PR
graph_TMFG <- tidygraph::tbl_graph(nodes = graph_TMFG$nodes, edges = graph_TMFG$edges, directed = FALSE) %>%
tidygraph::activate(nodes) %>%
mutate(centrality = tidygraph::centrality_degree())
as.list(graph_TMFG)$nodes %>%
group_by(dimension) %>%
arrange(desc(centrality)) %>%
slice_head(n=1) %>%
knitr::kable()| name | dimension | centrality |
|---|---|---|
| CVI | 1 | 20 |
| pNN20 | 2 | 32 |
| HFD | 3 | 12 |
| SI | 4 | 8 |
| LnHF | 5 | 16 |
| FuzzyEn | 6 | 26 |
Network Graph with Centrality
graph_glasso %>%
ggraph::ggraph(layout = "graphopt") +
ggraph::geom_edge_arc(aes(colour = link, width = link), strength = 0.05) +
ggraph::geom_node_point(aes(colour = dimension, size = centrality)) +
ggraph::geom_node_text(aes(label = name), fontface = "bold", repel = FALSE) +
ggraph::theme_graph() +
ggraph::scale_edge_width(range = c(0.2, 3), guide = "none") +
scale_color_material_d(palette = "rainbow", guide = "none") +
ggraph::scale_edge_color_gradientn(limits = c(0, 1), colours=c("#607D8B", "#263238"), guide = "none") +
scale_size_continuous(range = c(4, 20), guide = "none")
graph_TMFG %>%
ggraph::ggraph(layout = "graphopt") +
ggraph::geom_edge_arc(aes(colour = link, width = link), strength = 0.05) +
ggraph::geom_node_point(aes(colour = dimension, size = centrality)) +
ggraph::geom_node_text(aes(label = name), fontface = "bold", repel = FALSE) +
ggraph::theme_graph() +
ggraph::scale_edge_width(range = c(0.2, 3), guide = "none") +
scale_color_material_d(palette = "rainbow", guide = "none") +
ggraph::scale_edge_color_gradientn(limits = c(0, 1), colours=c("#607D8B", "#263238"), guide = "none") +
scale_size_continuous(range = c(4, 20), guide = "none")
Summary
Metaclustering Matrix
table <- table_clusters # [c("Index", "EGA", "EFA_3", "Mixture")]
# Initialize matrix
m <- matrix(data=NA, nrow=nrow(table), ncol=nrow(table), dimnames = list(table$Index, table$Index))
for(row in row.names(m)) {
for(col in colnames(m)) {
if(row == col) {
m[row, col] <- 0
next
}
subset <- table[table$Index %in% c(row, col), ]
rez <- sapply(subset[2:ncol(subset)], function(x) {
if(any(is.na(x))) {
NA
} else {
ifelse(length(unique(x[!is.na(x)])) == 1, 0, 1)
}
})
m[row, col] <- sum(rez, na.rm = TRUE) / length(na.omit(rez))
}
}m <- correlation::cor_sort(m, distance = "raw", hclust_method = "average")
# colnames(m) <- clean_names(colnames(m))
# rownames(m) <- clean_names(rownames(m))
# hrv_cols <- clean_names(hrv_cols)
# colnames(z) <- clean_names(colnames(z))
abs(m - 1) %>%
as.data.frame() %>%
rownames_to_column("Index1") %>%
datawizard::reshape_longer(cols = hrv_cols, colnames_to = "Index2", values_to = "Probability") %>%
mutate(Index1 = factor(Index1, levels = row.names(m)),
Index2 = factor(Index2, levels = rev(row.names(m)))) %>%
ggplot(aes(x = Index1, y = Index2)) +
geom_tile(aes(fill = Probability)) +
theme_minimal() +
theme(axis.text.x = element_text(angle = 90, hjust = 1)) +
labs(x = NULL, y = NULL) +
scale_fill_gradient2(low = "#2196F3", mid = "#9C27B0", high = "#FF5722", midpoint = 0.5) +
labs(title = "Metaclustering Heatmap", subtitle = "Probability of being grouped together accross multiple methods.")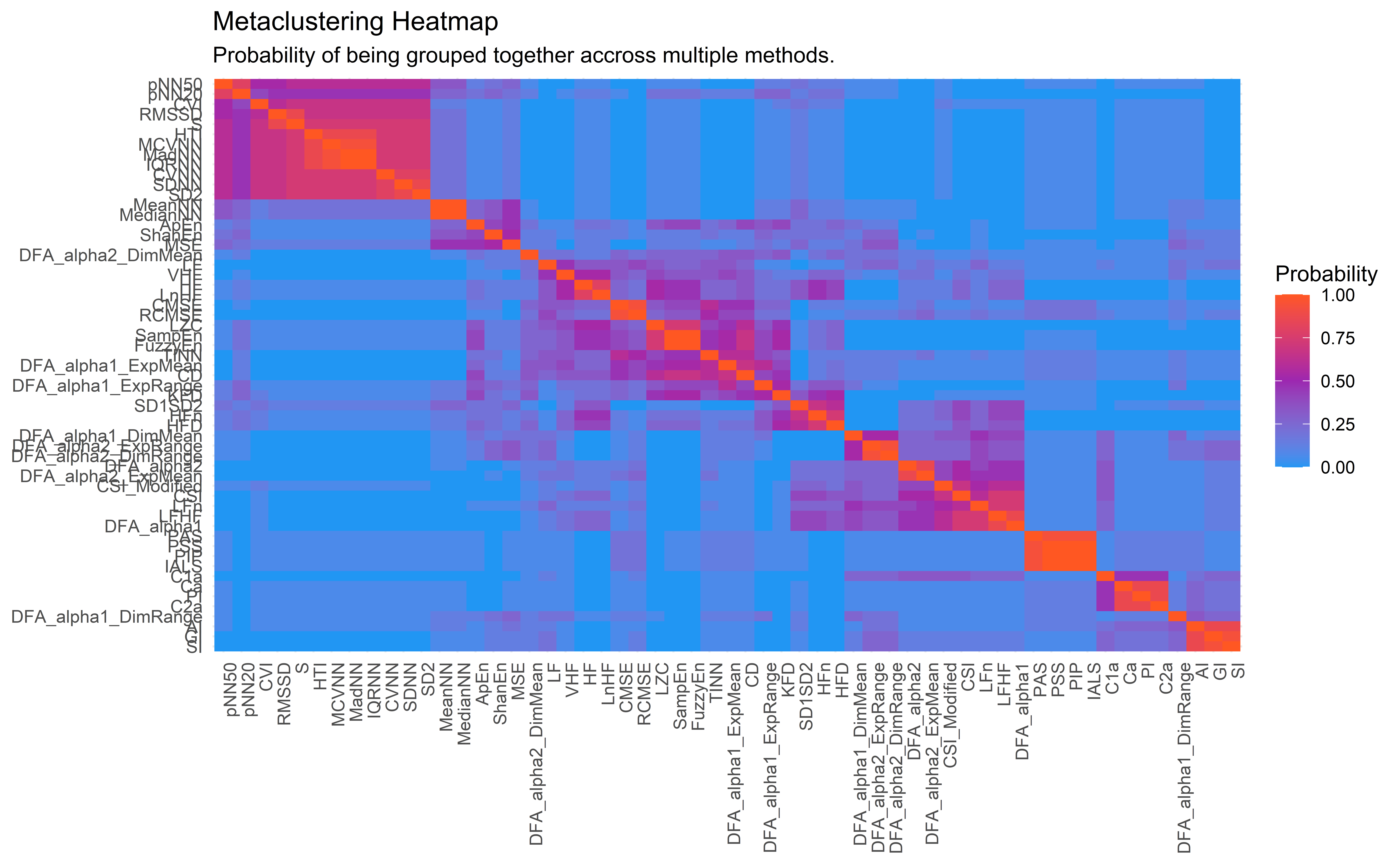
# ggsave("figures/figure_probability_matrix.png", width=12*0.9, height=9*0.9, dpi = 450)Hierarchical Structure of Metaclustering
model <- hclust(as.dist(m), method="average")
clusters <- cutree(model, h = 0.8)
clusters> pNN50 pNN20 CVI RMSSD
> 1 1 1 1
> S HTI MCVNN MadNN
> 1 1 1 1
> IQRNN CVNN SDNN SD2
> 1 1 1 1
> MeanNN MedianNN ApEn ShanEn
> 2 2 2 2
> MSE DFA_alpha2_DimMean LF VHF
> 2 3 3 3
> HF LnHF CMSE RCMSE
> 3 3 3 3
> LZC SampEn FuzzyEn TINN
> 3 3 3 3
> DFA_alpha1_ExpMean CD DFA_alpha1_ExpRange KFD
> 3 3 3 3
> SD1SD2 HFn HFD DFA_alpha1_DimMean
> 4 4 4 4
> DFA_alpha2_ExpRange DFA_alpha2_DimRange DFA_alpha2 DFA_alpha2_ExpMean
> 4 4 4 4
> CSI_Modified CSI LFn LFHF
> 4 4 4 4
> DFA_alpha1 PAS PSS PIP
> 4 5 5 5
> IALS C1a Ca PI
> 5 6 6 6
> C2a DFA_alpha1_DimRange AI GI
> 6 6 6 6
> SI
> 6# clusters2 <- c(
# pNN50 = 1, pNN20 = 1, CVI = 1, RMSSD = 1, S = 1, HTI = 1, MCVNN = 1,
# MadNN = 1, IQRNN = 1, CVNN = 1, SDNN = 1, SD2 = 1,
# MeanNN = 2, MedianNN = 2,
# ApEn = 2, ShanEn = 2, MSE = 2,
# DFA_alpha2_DimMean = 3,
# LF = 3, VHF = 3, HF = 3, LnHF = 3,
# CMSE = 3, RCMSE = 3,
# LZC = 3, SampEn = 3,
# FuzzyEn = 3, TINN = 3,
# DFA_alpha1_ExpMean = 3, CD = 3, DFA_alpha1_ExpRange = 3, KFD = 3,
# SD1SD2 = 4, HFn = 4, HFD = 4,
# DFA_alpha1_DimMean = 4, DFA_alpha2_ExpRange = 4, DFA_alpha2_DimRange = 4,
# DFA_alpha2 = 4, DFA_alpha2_ExpMean = 4, CSI_Modified = 4,
# CSI = 4, LFn = 4, LFHF = 4, DFA_alpha1 = 4,
# PAS = 5, PSS = 5, PIP = 5, IALS = 5,
# C1a = 6, Ca = 6, PI = 6, C2a = 6,
# DFA_alpha1_DimRange = 6,
# AI = 6, GI = 6, SI = 6)
# identify center of each cluster
dis <- parameters::cluster_centers(datawizard::data_transpose(z), clusters)
# reverse dis to have larger value for more central params
centrality <- datawizard::data_rescale(attributes(dis)$distance, to = c(max(attributes(dis)$distance), min(attributes(dis)$distance)))
names(centrality) <- hrv_cols
plot_dendrogram(model, clusters = clusters, ylim = -0.2, nudge_y = -0.03, angle = 90, points = TRUE, dis = centrality, h=0.8) +
# Lines to show "larger" clusters
# geom_line(data = data.frame(y = c(-0.02, -0.02), x = c(0, 5)), aes(x = x, y = y), size = 0.5, color = "#607D8B") +
# geom_line(data = data.frame(y = c(-0.02, -0.02), x = c(6, 15)), aes(x = x, y = y), size = 0.5, color = "#607D8B") +
# geom_line(data = data.frame(y = c(-0.02, -0.02), x = c(16, 22)), aes(x = x, y = y), size = 0.5, color = "#607D8B") +
# geom_line(data = data.frame(y = c(-0.02, -0.02), x = c(23, 34)), aes(x = x, y = y), size = 0.5, color = "#607D8B") +
# geom_line(data = data.frame(y = c(-0.02, -0.02), x = c(35, 42)), aes(x = x, y = y), size = 0.5, color = "#607D8B") +
theme(legend.position = "none")
# ggsave(file = "figures/meta-dendrogram.svg", plot=dendro, width=15, height=10)Metaclustering Network
Centrality
nodes <- data.frame(name = row.names(m),
Cluster = as.character(clusters))
row.names(nodes) <- NULL
edges <- m %>%
as.data.frame() %>%
datawizard::data_rename_rows(NULL) %>%
datawizard::data_rename(NULL) %>%
datawizard::reshape_longer(rows_to = "from",
names_to = "to",
values_to = "Probability") %>%
mutate(to = as.numeric(to)) %>%
filter(Probability > 0.8)
graph <- tidygraph::tbl_graph(nodes = nodes, edges = edges, directed = FALSE) %>%
tidygraph::activate(nodes) %>%
mutate(Centrality = tidygraph::centrality_degree())
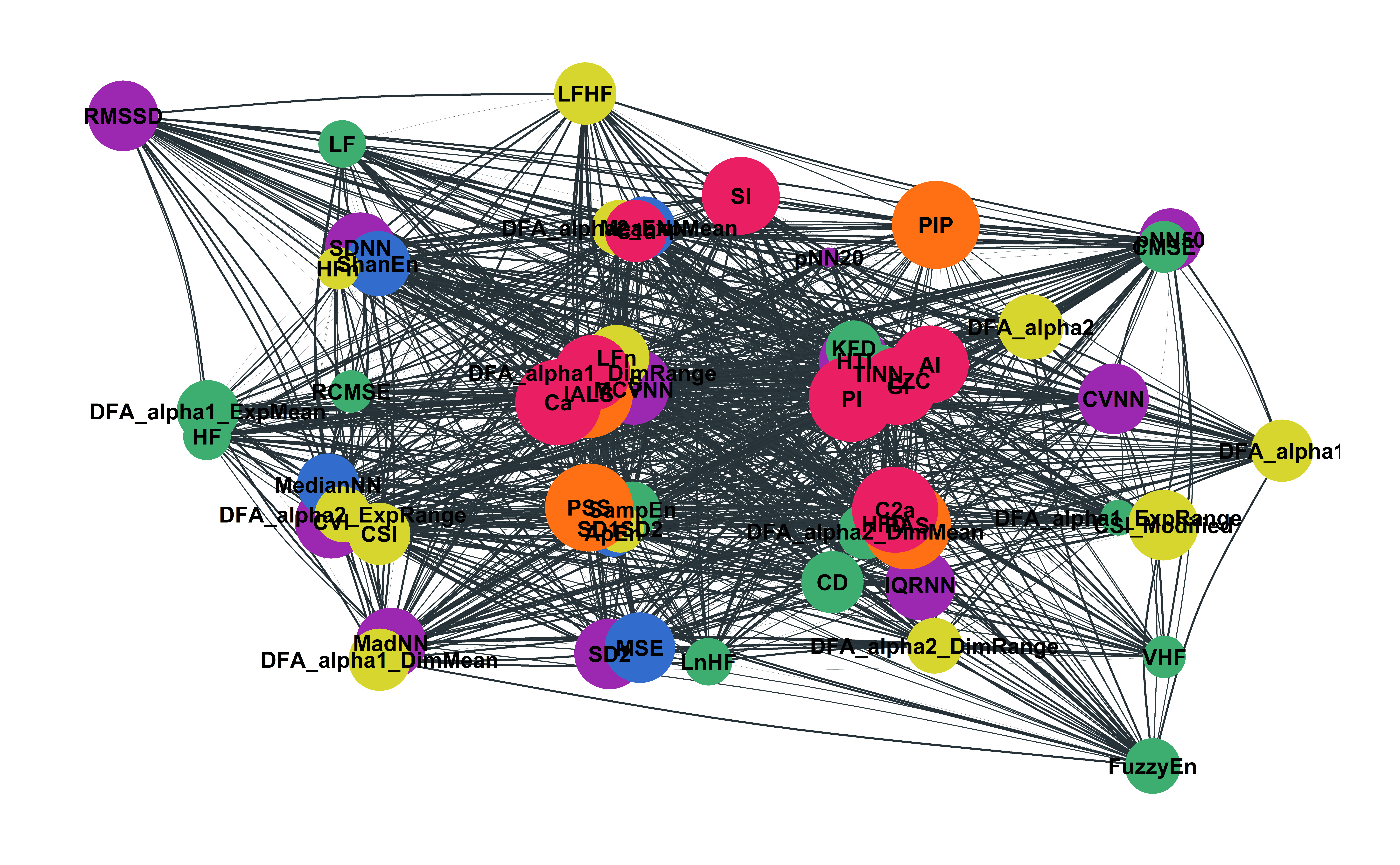
as.list(graph)$nodes %>%
group_by(Cluster) %>%
arrange(desc(Centrality)) %>%
knitr::kable()| name | Cluster | Centrality |
|---|---|---|
| PAS | 5 | 102 |
| PSS | 5 | 102 |
| PIP | 5 | 102 |
| IALS | 5 | 102 |
| Ca | 6 | 100 |
| PI | 6 | 100 |
| C2a | 6 | 100 |
| AI | 6 | 92 |
| GI | 6 | 92 |
| SI | 6 | 92 |
| DFA_alpha1_DimRange | 6 | 90 |
| CVI | 1 | 88 |
| RMSSD | 1 | 86 |
| S | 1 | 86 |
| HTI | 1 | 86 |
| MCVNN | 1 | 86 |
| MadNN | 1 | 86 |
| IQRNN | 1 | 86 |
| CVNN | 1 | 86 |
| SDNN | 1 | 86 |
| SD2 | 1 | 86 |
| MSE | 2 | 86 |
| CSI_Modified | 4 | 86 |
| ShanEn | 2 | 82 |
| DFA_alpha2 | 4 | 82 |
| LFn | 4 | 82 |
| pNN50 | 1 | 80 |
| MeanNN | 2 | 80 |
| MedianNN | 2 | 80 |
| DFA_alpha1_ExpMean | 3 | 80 |
| CD | 3 | 80 |
| DFA_alpha1_DimMean | 4 | 80 |
| CSI | 4 | 80 |
| LFHF | 4 | 80 |
| DFA_alpha1 | 4 | 80 |
| C1a | 6 | 80 |
| TINN | 3 | 78 |
| DFA_alpha2_DimMean | 3 | 76 |
| LZC | 3 | 76 |
| SampEn | 3 | 76 |
| FuzzyEn | 3 | 76 |
| KFD | 3 | 76 |
| DFA_alpha2_ExpRange | 4 | 76 |
| DFA_alpha2_DimRange | 4 | 76 |
| DFA_alpha2_ExpMean | 4 | 76 |
| CMSE | 3 | 74 |
| ApEn | 2 | 72 |
| LF | 3 | 72 |
| HF | 3 | 72 |
| LnHF | 3 | 72 |
| SD1SD2 | 4 | 72 |
| VHF | 3 | 70 |
| RCMSE | 3 | 70 |
| HFn | 4 | 70 |
| DFA_alpha1_ExpRange | 3 | 68 |
| pNN20 | 1 | 66 |
| HFD | 4 | 66 |
Network Graph
graph %>%
ggraph::ggraph(layout = "graphopt") +
geom_edge_arc(aes(colour = Probability, width = Probability), strength = 0.05) +
geom_node_point(aes(colour = Cluster, size = Centrality)) +
geom_node_text(aes(label = name), fontface = "bold", repel = FALSE) +
ggraph::theme_graph() +
scale_edge_width(range = c(0.05, 0.5), guide = "none") +
scale_color_material_d(palette = "rainbow", guide = "none") +
scale_edge_color_gradientn(limits = c(0, 1), colours=c("#607D8B", "#263238"), guide = "none") +
scale_size_continuous(range = c(4, 20), guide = "none")